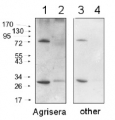
1

Anti-Lhcb5 | CP26 (Lhcb5) homolog, Chlamydomonas
AS09 407 | Clonality: Polyclonal | Host: Rabbit | Reactivity: Chlamydomonas reinhardtii
- Product Info
-
Immunogen: KLH-conjugated synthetic peptide derived from Chlamydomonas reinhardtii Lhcb5 protein sequence, UniProt:Q9FEK6
Host: Rabbit Clonality: Polyclonal Purity: Serum Format: Lyophilized Quantity: 50 µl Reconstitution: For reconstitution add 50 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Western blot (WB) Recommended dilution: 1 : 5000-1 : 10 000 (WB) Expected | apparent MW: 30 | 26 kDa
- Reactivity
-
Confirmed reactivity: Chlamydomonas reinhardtii Predicted reactivity: Chlamydomonas reinhardtii
Not reactive in: Other algae - Application Examples
-
Application example 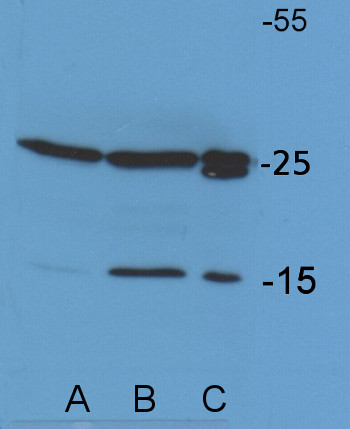
Chlamydomonas reinhardtii membrane extract (A), Chlamydomonas reinhardtii total cell extract, prepared by sonication, loading 14 µl equivalent to 30 µg of total protein (B), Chlamydomonas reinhardtii total cell extract, rapid, prepared directly by spinning down the cells and lysis of cell pellet in SDS-PAGE sample buffer and loading 14 µl equivalent to 98 µg of total protein (C), denatured at 100°C for 5 min. were separated on 15 % SDS-PAGE and blotted 1h to PVDF using tank transfer. Blots were blocked with 1 % blocking buffer (2 ml blocking reagent stock solution ROCHE 11 520 709 001 in 20 ml TBS) for ON at 4°C without agitation. Blot was incubated in the primary antibody at a dilution of 1: 25 000 for 1h at RT with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, then washed once for 15 min and 3 times for 5 min in TBS-T at RT with agitation. Blot was incubated in secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated, from Agrisera AS09 602) diluted to 1:20 000 in for 1h at RT with agitation. The blot was washed as above and developed according to manufacture instructions. Exposure time was 8 seconds.
Courtesy of Nadine Coosemans, Laboratoire de génétique et physiologie des microalgues, Université de Liège, BelgiumApplication examples: 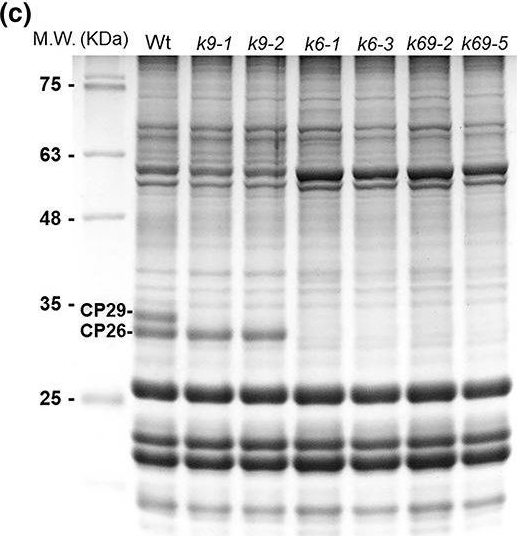
Reactant: Chlamydomonas reinhardtii (Green Alga)
Application: Western Blotting
Pudmed ID: 31724187
Journal: Plant Cell Environ
Figure Number: 1C
Published Date: 2020-02-01
First Author: Cazzaniga, S., Kim, M., et al.
Impact Factor: 6.599
Open PublicationCP26? and CP29?targeted gene disruption in Chlamydomonas reinhardtii by CRISPR/Cas9 genome editing. (a) DNA sequence alignment of wild type (Wt) and mutants on cp26 (k6) and cp29 (k9) obtained by CRISPR/Cas9 genome editing. The 20?bp target sequence of sgRNA is reported before the red?coloured PAM sequence. In the case of k9 mutant, insertions (k9?1) or deletions were induced (k9?2) by non?homologous repair, whereas in the case of k6 mutant, a Cas9?mediated hygromycin resistance cassette insertion was obtained in the target site. Double?mutant k69 was obtained by hygromycin resistance cassette insertion in a k9 background. (b) PCR amplification of cp26 and cp29 CDS sequences from Wt, k6, k9, and k69 cDNA. In the case of k6 and k69 mutants, the cp26 CDS cannot be amplified due to hygromycin cassette insertion. (c) SDS?PAGE analysis of Wt and mutant strains performed with the Tris?Tricine buffer system. Fifteen microgram of Chl was loaded in each lane. Molecular weight (MW) markers are indicated on the left. CP26 and CP29 bands are marked. (d) Immunoblot with specific antibodies directed against CP26 and CP29 on the same lanes of (c). Immunoblot against CP43 was added as control of the loading. (e) qRT?PCR on cp26 and cp29 genes in Wt and mutant strains. rack1 gene was used as control for qRT?PCR (see details in Figure S3)
- Additional Information
-
Additional information (application): For optimal detection, load/well should be from 5 to 10 µg of total protein. Higher protein load combined with weak blocking will contribute to detection of other bands. - Background
-
Background: Lhcb5 is one of the 3 minor, highly conserved chlorophyll a/b-binding proteins associated with Photosystem II in plants and algae. As a part of the inner light-harvesting antenna it has been sugested to regulate (together with Lhcb4 and Lhcb6) the energy flow from the outer LHCII antenna to the PSII reaction center.
- Product Citations
-
Selected references: Cazzaniga et al. (2022). Engineering astaxanthin accumulation reduces photoinhibition and increases biomass productivity under high light in Chlamydomonas reinhardtii. Biotechnol Biofuels Bioprod. 2022 Jul 11;15(1):77. doi: 10.1186/s13068-022-02173-3. PMID: 35820961; PMCID: PMC9277849.
Cecchin et al (2021) LPA2 protein is involved in photosystem II assembly in Chlamydomonas reinhardtii. Plant J. 2021 Jul 4. doi: 10.1111/tpj.15405. Epub ahead of print. PMID: 34218480.
Gonzaga Heredia-Martinez et al. (2018). Chloroplast damage induced by the inhibition of fatty acid synthesis triggers autophagy in Chlamydomonas. Plant Physiol, Sept. 2018.
Correa-Galvis et al. (2016). Photosystem II Subunit PsbS Is Involved in the Induction of LHCSR Protein-dependent Energy Dissipation in Chlamydomonas reinhardtii. J Biol Chem. 2016 Aug 12;291(33):17478-87. doi: 10.1074/jbc.M116.737312.
Muranaka et al. (2015). TEF30 interacts with photosystem II monomers and is involved in the repair of photodamaged photosystem II in Chlamydomonas reinhardtii. Plant Physiol. 2015 Dec 7. pii: pp.01458.2015.
Drop et. al (2014). Consequences of state transitions on the structural and functional organization of Photosystem I in the green alga Chlamydomonas reinhardtii. Plant J. 2014 Feb 8. doi: 10.1111/tpj.12459.
Takahashi et al. (2006). Identification of the mobile light-harvesting complex II polypeptides for state transitions in Chlamydomonas reinhardtii. PNAS 103:477-482 - Protocols
-
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extracts
Oxygenic photosynthesis poster by prof. Govindjee and Dr. Shevela
Z-scheme of photosynthetic electron transport by prof. Govindjee and Dr. Björn and Dr. Shevela - Reviews:
-
This product doesn't have any reviews.