1
Anti-AGO1 | Argonaute 1
AS09 527 | Clonality: Polyclonal | Host: Rabbit | Reactivity: Arabidopsis thaliana, Nicotiana benthamiana
- Product Info
-
Immunogen: KLH-conjugated, N-terminal peptide of Arabidopsis thaliana AGO1 O04379, At1g48410
Host: Rabbit Clonality: Polyclonal Purity: Immunogen affinity purified serum in PBS pH 7.4. Format: Lyophilized Quantity: 50 µg Reconstitution: For reconstitution add 50 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Chromatin Immunoprecipitation (ChIP), Immunofluorescence (IF), Immunolocalization (IL), small-RNA-IP-Seq, Western blot (WB) Recommended dilution: 2 µg (ChIP), 1: 200 (IF), (IL), small-RNA-IP-Seq, 1 : 5000-1 : 10 000 (WB) Expected | apparent MW: 116.4 | 130 kDa - Reactivity
-
Confirmed reactivity: Arabidopsis thaliana, Hyacinthus orientalis, Nicotiana benthamiana
Predicted reactivity: Brassica napus, Brassica pekinensis, Capsella rubella, Cucumis sativus, Glycine max, Malus domestica, Pisum sativum, Ricinus communis, Solanum lycopersicum, Solanum tuberosum, Vitis vinifera
Species of your interest not listed? Contact usNot reactive in: Chlamydomonas reinhardtii, Medicago sativa, Salvia sp., Spinacia oleracea, Oryza sativa, Phaseolus vulgaris, Triticum aestivum, Zea mays
Mammals - Application Examples
-
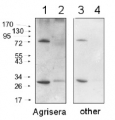
80 µg of Arabidopsis thaliana soluble total cell extract (extracted in 20 mMTris pH 7.5, 5mM MgCl2, 2.5mM DTT, 300 mM NaCl, 0.1% NP-40, 1% proteasome inhibitor MG132) was separated on 6% SDS-PAGE and blotted 1h to nitrocellulose. Filters were blocked 1h with 5% low-fat milk powder in TBS-TT (0.25% TWEEN20; 0.1% Triton-X) and probed with anti-AGO1 antibody (1:10 000, 1h) and secondary anti-rabbit (1:10000, 1 h) antibody (HRP conjugated) in TBS-TT containing 5% low fat milk powder. Antibody incubations were followed by washings in TBS-TT. All steps were performed at RT with agitation. Blots were developed for 5 min with ECL-PLUS detection reagent according to the manufacturer's instructions. Exposure time was 5 seconds.
Roche's protease inhibitor cocktail (no EDTA) can also be applied in extraction buffer.Courtesy Dr. Ericka Havecker, University of Cambridge, UK
Application examples: 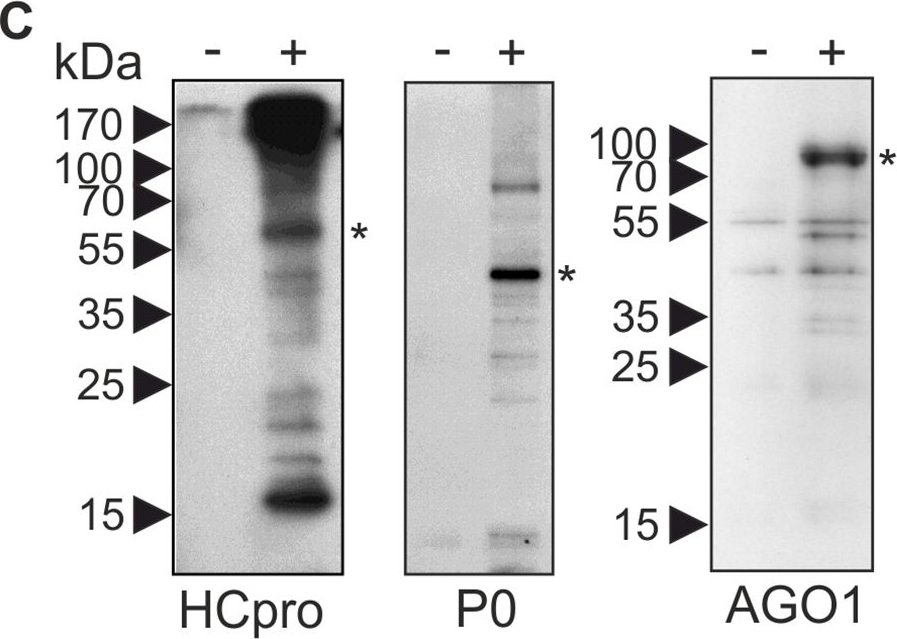
Reactant: Nicotiana benthamiana
Application: Western Blotting
Pudmed ID: 26641460
Journal: PLoS Pathog
Figure Number: 3C
Published Date: 2015-12-01
First Author: Hafrén, A., Lõhmus, A., et al.
Impact Factor: 6.506
Open PublicationPGs are RNA granules to which also PVA RNA can localize.(A) Fractions containing PGs labeled with P0YFP were isolated from PVACPmut infected leaf lysates (+ sample), as described in materials and methods. Lysates from non-infected leaves expressing P0YFP were fractionated similarly (- sample). Three parallel fractions of each sample type were used for fluorescence quantification and relative fluorescence units are given as a mean ± standard deviation. (B) Fractions quantified in (A) were imaged using epifluorescence microscopy to show successful capture of PVA-induced PGs. Scale bar; 500 ?m. (C) PG fractions were isolated from mock-infiltrated leaves (-) and PVACPmut RNA-expressing leaves (+) at 3 DAI and subjected to a western blot detection of endogenous HCpro, P0 and AGO1. The asterisks denote the expected position of the corresponding protein. (D) Similar samples enriched for PGs as in (A and B), were incubated with or without RNAse A and imaged using epifluorescence microscopy. Scale bar; 20 ?m. (E) RNAse A-mediated release of fluorescence from isolated PGs (D) was quantified by analyzing fluorescence in total, soluble and low-speed pellet fractions after RNAse A treatment compared to control samples (n = 3). (F) PG fractions were prepared from leaves expressing PVA together with either P0YFP (control) or Strep-III-tagged P0 (P0SIII), and subjected to Strep-tag based affinity purification. RNA was isolated from the affinity-purified samples and subjected to reverse transcription (RT+), followed by PCR detection of viral RNA. Total RNA from PVA infected leaves was used as a positive control for RT-PCR. The RT-minus control (RT-) was negative. (G) Bacteriophage ? B-box RNA elements were fused to the 3´ UTR within PVA?GDD icDNA (PVAB-box; S1 Fig). Binding of ?N22RFP to the B-box RNA element enabled visualization of PVAB-box RNA in vivo. PGs were induced either by PVA?GDD (control) or PVAB-box and visualized by P0YFP (green channel), and ?N22RFP was co-expressed to label B-box RNA. The RFP signal was mainly found in nuclei due to the nuclear localization signal present in ?N22RFP, but also in the cytoplasm and PGs in the presence of PVAB-box (magenta channel). The images are projections of Z-stacks with a single layer inset from the area indicated with an arrow. Scale bar; 10 ?m.
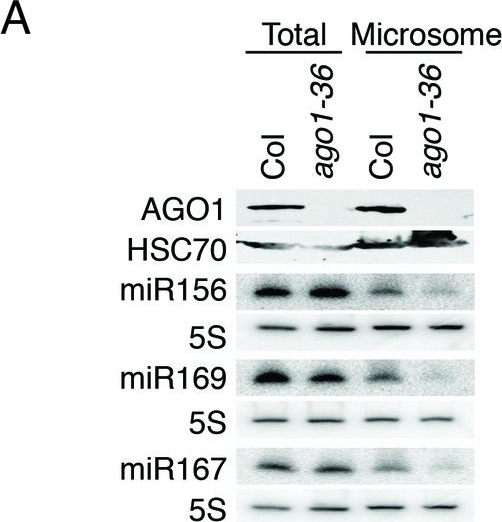
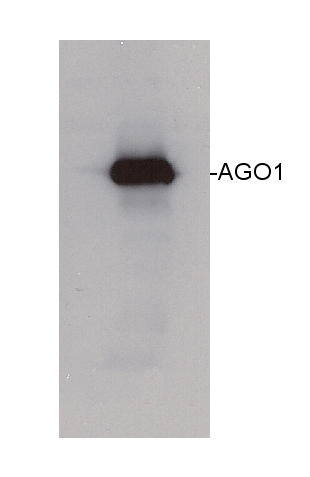
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 27938667
Journal: Elife
Figure Number: 5A
Published Date: 2016-12-12
First Author: Li, S., Le, B., et al.
Impact Factor: 7.448
Open PublicationmiRNAs and ta-siRNAs are recruited to membranes by AGO1.(A) Detection of three miRNAs in total extracts and the microsomal fraction in wild type (Col) and ago1-36. The ago1-36 mutant lacks the full-length AGO1 protein as shown by western blotting. HSC70 was a loading control. The three miRNAs were detected by northern blotting. 5S rRNA was an internal control and shown below each miRNA blot. The microsomal levels of the miRNAs were reduced in ago1-36. (B) A scatter plot showing comparable miRNA abundance in wild type (WT) and ago1-27 total extracts. Inset: box plots illustrating the distribution of miRNA RPMR values in WT and ago1-27. (C) A scatter plot showing that miRNAs have reduced microsomal enrichment in ago1-27 as compared with wild type. Microsomal enrichment was measured by M/T (ratio of levels in microsome vs. those in total extract). The degree of microsomal enrichment of miR390 (yellow dots) is largely unchanged, while that of most known ta-siRNA/phasiRNA triggers (red dots) is decreased in ago1-27. miR173’s (green dot) microsomal enrichment was weakly affected. (D) A scatter plot showing that 21-nt and 22-nt ta-siRNAs from TAS1-4 loci have reduced microsomal enrichment in ago1-27 as compared with wild type.DOI:http://dx.doi.org/10.7554/eLife.22750.010
- Additional Information
-
Additional information: Antibody binds microRNA and tasiRNAs, preference for 21nt miRNAs with 5'U.
To detect AGO1 in Nicotiana benthamiana, please inquire.
Recommended for detection of AGO1: extreme low femtogram range chemiluminescent detection reagentAdditional information (application): AGO expression may be tissue specific and using floral tissue is recommended where most of the AGOs are expressed the highest. Use of proteasome inhibitors as MG132 can help to stabilize AGO proteins during extraction procedure. Buffer for extraction of AGO proteins: Paudel et al. 2018.The AGO1 antibody is extremely specific to AGO1 and does not cross-react with other antibodies. The evidence is 1) the peptide to which it was raised is at the very N-terminus of the protein and is not present in other AGOs 2) aAGO1 does not cross react with the AGOs which are overexpressed (AGO2, AGO3, AGO4, AGO5, AGO6, AGO9) using a western blot.
TCA acetone precipitation method - Background
-
Background: AGO1 belongs to a group of argonaute proteins which are catalytic component of the RNA-incudes silencing complex (RISC). This protein complex is responsible for the gene silencing (RNAi).
- Product Citations
-
Selected references: Dalmadi et al. (2025). Remote precursor elements can modulate RNA induced silencing complex-loading efficiency of miR168 in Arabidopsis. Plant J . 2025 May;122(3):e70195. doi: 10.1111/tpj.70195.
Bressendorff et al. (2025). Importance of an N-terminal structural switch in the distinction between small RNA-bound and free ARGONAUTE. Nat Struct Mol Biol. 2025 Jan 7. doi: 10.1038/s41594-024-01446-9.
Nasfi et al. (2024). A pipeline for validation of Serendipita indica effector-like sRNA suggests cross-kingdom communication in the symbiosis with Arabidopsis. J Exp Bot . 2024 Dec 26:erae515. doi: 10.1093/jxb/erae515
Li et al. (2024). RNA helicase Brr2a promotes miRNA biogenesis by properly remodelling secondary structure of pri-miRNAs. Nat Plants . 2024 Oct;10(10):1532-1547. doi: 10.1038/s41477-024-01788-8.
Mirlohi et al. (2024). An AGO10: miR165/6 module regulates meristem activity and xylem development in the Arabidopsis root. EMBO J. 2024 May;43(9):1843-1869.doi: 10.1038/s44318-024-00071-y.
Martín-Merchán et al. (2024). Arabidopsis AGO1 N-terminal extension acts as an essential hub for PRMT5 interaction and post-translational modifications. Nucleic Acids Res . 2024 May 20:gkae387.doi: 10.1093/nar/gkae387.
Barre-Villeneuve et al. (2024). The unique dual targeting of AGO1 by two types of PRMT enzymes promotes phasiRNA loading in Arabidopsis thaliana. Nucleic Acids Res. 2024 Mar 21;52(5):2480-2497. doi: 10.1093/nar/gkae045.
Blagojevic et al. (2024). Heat stress promotes Arabidopsis AGO1 phase separation and association with stress granule components. iScience. 2024 Feb 6;27(3):109151. doi: 10.1016/j.isci.2024.109151. eCollection 2024 Mar 15.
Bradamante et al. (2023). Two ARG ONAUTE proteins loaded with transposon-derived small RNAs are associated with the reproductive cell lineage in Arabidopsis. Plant Cell. 2023 Dec 7:koad295.doi: 10.1093/plcell/koad295.
Liang et al. (2022). Arabidopsis RBV is a conserved WD40 repeat protein that promotes microRNA biogenesis and ARGONAUTE1 loading. Nat Commun. 2022 Mar 8;13(1):1217. doi: 10.1038/s41467-022-28872-x. PMID: 35260568; PMCID: PMC8904849.
Hacquard et al. (2022) The Arabidopsis F-box protein FBW2 targets AGO1 for degradation to prevent spurious loading of illegitimate small RNA. Cell Rep. 2022 Apr 12;39(2):110671. doi: 10.1016/j.celrep.2022.110671. PMID: 35417704; PMCID: PMC9035678.
Oliver et al. (2022) The miRNome function transitions from regulating developmental genes to transposable elements during pollen maturation. Plant Cell. 2022 Feb 3;34(2):784-801. doi: 10.1093/plcell/koab280. PMID: 34755870; PMCID: PMC8824631.
Lakatos et al. (2022). In Arabidopsis thaliana, RNA-Induced Silencing Complex-Loading of MicroRNAs Plays a Minor Regulatory Role During Photomorphogenesis Except for miR163. Front Plant Sci. 2022 Jul 13;13:854869. doi: 10.3389/fpls.2022.854869. PMID: 35909792; PMCID: PMC9326452.
Meng et al. (2022) The novel activity of Argonautes in intron splicing: A transcriptome-wide survey in plants. J Plant Physiol. 2022 Jan 31;270:153632. doi: 10.1016/j.jplph.2022.153632. Epub ahead of print. PMID: 35114616.
Cabezas-Fuster et al. (2022). Missplicing suppressor alleles of Arabidopsis PRE-MRNA PROCESSING FACTOR 8 increase splicing fidelity by reducing the use of novel splice sites. Nucleic Acids Res. 2022 Jun 10;50(10):5513-5527. doi: 10.1093/nar/gkac338. PMID: 35639749; PMCID: PMC9177961.
Delenko et al. (2022) MicroRNA biogenesis and activity in plant cell dedifferentiation stimulated by cell wall removal. BMC Plant Biol. 2022 Jan 3;22(1):9. doi: 10.1186/s12870-021-03323-9. PMID: 34979922; PMCID: PMC8722089. (immunofluorescence)
Oliver & Martinez. (2021) Accumulation dynamics of ARGONAUTE proteins during meiosis in Arabidopsis. Plant Reprod. 2021 Nov 23. doi: 10.1007/s00497-021-00434-z. Epub ahead of print. PMID: 34812935.
Dalmadi et al. (2021) Controlled RISC loading efficiency of miR168 defined by miRNA duplex structure adjusts ARGONAUTE1 homeostasis. Nucleic Acids Res. 2021 Dec 16;49(22):12912-12928. doi: 10.1093/nar/gkab1138. PMID: 34850097; PMCID: PMC8682782.
Clavel et al. (2021) Atypical molecular features of RNA silencing against the phloem-restricted polerovirus TuYV. Nucleic Acids Res. 2021 Nov 8;49(19):11274-11293. doi: 10.1093/nar/gkab802. PMID: 34614168; PMCID: PMC8565345.
Brioudes et al. (2021) HASTY, the Arabidopsis EXPORTIN5 ortholog, regulates cell-to-cell and vascular microRNA movement. EMBO J. 2021 Aug 2;40(15):e107455. doi: 10.15252/embj.2020107455. Epub 2021 Jun 21. PMID: 34152631; PMCID: PMC8327949. (Immunoprecipitation)
Schwenk et al. (2021) Uncovering a novel function of the CCR4-NOT complex in phytochrome A-mediated light signalling in plants. Elife. 2021 Mar 30;10:e63697. doi: 10.7554/eLife.63697. PMID: 33783355; PMCID: PMC8009681.
Dunker, Lederer, and Weiberg. (2021). Plant ARGONAUTE Protein Immunopurification for Pathogen Cross Kingdom Small RNA Analysis. Bio-protocol 11(3): e3911. DOI: 10.21769/BioProtoc.3911.
Dunker et al. (2020). Oomycete small RNAs bind to the plant RNA-induced silencing complex for virulence. Elife. 2020 May 22;9:e56096.doi: 10.7554/eLife.56096.
Niedojadlo et al. (2020). Dynamic distribution of ARGONAUTE1 (AGO1) and ARGONAUTE4 (AGO4) in Hyacinthus orientalis L. pollen grains and pollen tubes growing in vitro. Protoplasma. 2020 Jan 8. doi: 10.1007/s00709-019-01463-2.
You et al. (2019). FIERY1 promotes microRNA accumulation by suppressing rRNA-derived small interfering RNAs in Arabidopsis. Nat Commun. 2019 Sep 27;10(1):4424. doi: 10.1038/s41467-019-12379-z
Dalmadi et al. (2019). AGO-unbound cytosolic pool of mature miRNAs in plant cells reveals a novel regulatory step at AGO1 loading. Nucleic Acids Res. 2019 Aug 8. pii: gkz690. doi: 10.1093/nar/gkz690.
Li (2019). The Isolation of Total and Membrane-Bound Polysomes from Arabidopsis and the Detection of Their Associated AGO1 and sRNAs. Methods Mol Biol. 2019;1932:317-333. doi: 10.1007/978-1-4939-9042-9_23.
Sprunck et al. (2019). Elucidating small RNA pathways in Arabidopsis thaliana egg cells. doi: 10.1101/525956
Chen et al. (2018). Structural and biochemical insights into small RNA 3' end trimming by Arabidopsis SDN1. Nat Commun. 2018 Sep 4;9(1):3585. doi: 10.1038/s41467-018-05942-7. (Immunoprecipitation)
Bologna et al. (2018). Nucleo-cytosolic Shuttling of ARGONAUTE1 Prompts a Revised Model of the Plant MicroRNA Pathway. Mol Cell. 2018 Feb 15;69(4):709-719.e5. doi: 10.1016/j.molcel.2018.01.007.
Zhang et al. (2017). RISC-interacting clearing 3'- 5' exoribonucleases (RICEs) degrade uridylated cleavage fragments to maintain functional RISC in Arabidopsis thaliana. Elife. 2017 May 2;6. pii: e24466. doi: 10.7554/eLife.24466.
Schalk et al. (2017). Small RNA-mediated repair of UV-induced DNA lesions by the DNA DAMAGE-BINDING PROTEIN 2 and ARGONAUTE 1. Proc Natl Acad Sci U S A. 2017 Mar 21. pii: 201618834. doi: 10.1073/pnas.1618834114.
Dolata et al. (2016). Salt Stress Reveals a New Role for ARGONAUTE1 in miRNA Biogenesis at the Transcriptional and Posttranscriptional Levels. Plant Physiol. 2016 Sep;172(1):297-312. doi: 10.1104/pp.16.00830. (ChIP and Immunolocalization)
Pumplin et al. (2016). DNA Methylation Influences the Expression of DICER-LIKE4 Isoforms, Which Encode Proteins of Alternative Localization and Function. The Plant Cell November 14, 2016 tpc.00554.2016 . Advance Publication November 14, 2016.
Minoia et al. (2014). Specific Argonautes Selectively Bind Small RNAs Derived from Potato Spindle Tuber Viroid and Attenuate Viroid Accumulation In Vivo. J Virol. 2014 Oct 15;88(20):11933-45. doi: 10.1128/JVI.01404-14. Epub 2014 Aug 6.
Haveckeret al. (2010). The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell. 2010 Feb;22(2):321-34. doi: 10.1105/tpc.109.072199. (small-RNA-IP-Seq) - Protocols
-
TCA/Acetone protein precipitation method for plant proteins
- Grind plant tissue in a liquid nitrogen.
- Continue grinding with 10% TCA solution in acetone (ice cold).
- Precipitate overnight in -20C.
- Spin at 4°C for 1min, 17k rpm > wash with ice cold acetone until you obtain a white pellet.
- Dissolve the pellet in buffer of choice (for example 8M urea containing 5mM DTT, or denaturate in SDS protein loading buffer for 10 min. at 70°C)
- Clarify supernatant
- Measure protein concentration.
- Proceed with a western blot.
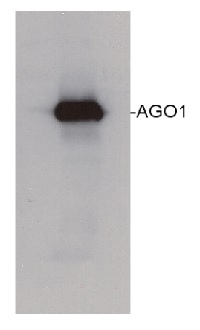
Example of a western blot result obtained with this method, which allows high protein load per well, can be found below.
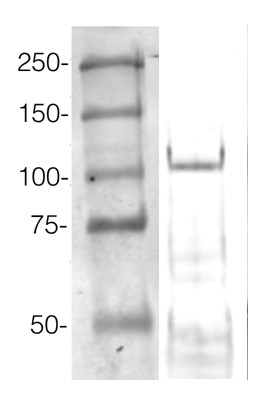
360 µg/well of Arabidopsis thaliana protein extracted by TCA-acetone precipitation from floral tissue and saturated in 8M urea were separated on 15% SDS-PAGE and blotted for 1hour to 0.2 µm nitrocellulose at 100V using wet transfer system. Blots were blocked with 0.5% cold fish gelatin for 1hr at room temp with agitation. Blot was incubated in the primary antibody at a dilution of 1:2500 for an hour at RT with agitation. The blots were washed with 3X 15min TBS-TT at RT with agitation. Blots as incubated in the secondary antibody (DayLight 800) 1:5000 dilution for 30 min. at RT with agitation and washed 1X with TBSTT for 15 min, 1X with TBST for 15min before scanning with the ODyssey IRD scanner.
Courtesy of Dr. Betty Chung and Pawel Baster, University of Cambridge, United Kingdom
BACK to Plant Protocols page - Reviews:
-
Manfred Heinlein | 2025-06-24This antibody against AGO1 performs exceptionally well and is highly sensitive for both western blot and co-immunoprecipitation applications. It provides clear and specific signal with minimal background, making it a reliable and efficient tool for protein detection and interaction studies. Highly recommended.Ágnes Dalmadi | 2021-12-08We have used it in two NAR publications for WB and IP. It gave strong and specific signal in case of every tissue type.Jacinthe AZEVEDO FAVORY | 2021-10-26Very good for western blots and IPsXiuren Zhang | 2013-03-11This antibody is excellent for both IP and western blot.Xiuren Zhang | 2013-03-11This antibody is excellent for both IP and western blot.Katarzyna Niedojadło | 2013-02-27I checked by immunocytochemistry but I have problems with Western blotting.Katarzyna NiedojadÅ‚o | 2012-04-23Antibodies were used in immynocytochemistry methods in Hyacinthus orientalis and diluted 1:100.
Accessories
This set contains:
- 4 Anti-AGO antibodies of your choice, chosen from a drop down menu below
- Matching secondary antibody, HRP conjugated, min. 1: 25 000, 1h/RT (AS09 602 - trial)