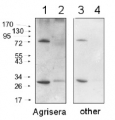
1
Anti-COXII | Plant Cytochrome oxidase subunit II (affinity purified)
AS04 053A | Clonality: Polyclonal | Host: Rabbit | Reactivity: Eudicots, monocots, Physcomitrella patens | Cellular [compartment marker] of mitochondrial inner membrane
- Product Info
-
Immunogen: KLH-conugated synthetic peptide fully conserved in all available protein sequences from eudicots including Arabidopsis thaliana AtmG00160, monocots including Oryza sativa P04373 and Physcomitrella patens Q1XGA9
Host: Rabbit Clonality: Polyclonal Purity: Immunogen affinity purified serum in PBS pH 7.4. Format: Lyophilized Quantity: 50 µg Reconstitution: For reconstitution add 50 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Blue Native-PAGE (BN-PAGE), Western blot (WB) Recommended dilution: 1 : 1000 (BN-PAGE), 1 : 1000 (WB) Expected | apparent MW: 29.4 | 30 kDa (for Arabidopsis thaliana) - Reactivity
-
Confirmed reactivity: Actinidia chinensis, Arabidopsis thaliana (leaf extract and isolated mitochondria), Betula nana, Brassica napus, Brassica oleracea, Cicer aretinum, Cucumis melo, Cucumis sativus, Erophorum vaginatum, Hordeum vulgare, Lilium longifolorum, Nicotiana tabacum, Picea abies, Plantago major, Plantago euryphylla, Silene uniflora, Silene dioica, Physcomitrium patens, Triticum aestivum, Triticum durum Desf., Zea mays, Vicia faba, Quercus rubra
Predicted reactivity: Cucumis melo, Glycine max, Gossypium hirsutum, Neurachne alopecuroidea, Neurachne minor, Neurachne muelleri, Oryza sativa, Physcomitrium patens, Pisum sativum, Triticum aestivum, Vigna radiata
Species of your interest not listed? Contact usNot reactive in: Mangifera indica, Saccharina japonica - Application Examples
-
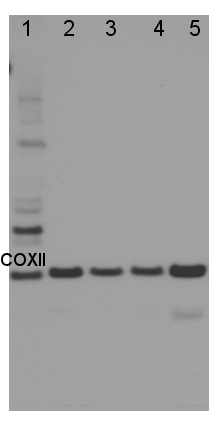
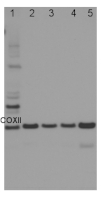
1.8 µg of total protein from (1) Arabidopsis thaliana leaf , (2) Plantago major leaf, (3) Plantago europhylla leaf, (4) Silene dioica leaf, (5) Silene uniflora leaf were separated on 4-12% NuPage (Invitrogen) LDS-PAGE and blotted 1h to PVDF. Primary antibody was used in 1: 1000 dilution. Detection was performed using chemiluminescence, following manufacture's recommendations.
Application examples: 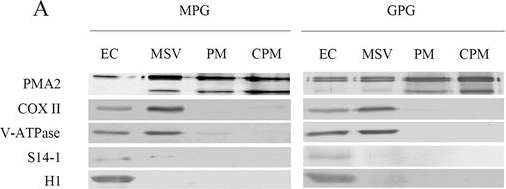
Reactant: Oryza sativa (Asian rice)
Application: Western Blotting
Pudmed ID: 28056797
Journal: BMC Plant Biol
Figure Number: 2A
Published Date: 2017-01-05
First Author: Yang, N. & Wang, T.
Impact Factor: 4.142
Open PublicationPlasma membrane (PM) purity verification and PM-related proteins evaluation. a Western blot examination of plasma membrane enrichment. Proteins from entire cell (EC) lysates, microsomal vesicles (MSV), PM (plasma membrane vesicles) and carbonate-washed PM (CPM) were separated by 10% SDS-PAGE, transferred to PVDF membranes and detected with antibodies for PMA2, a PM marker; COXII, a mitochondrial marker; V-ATPase, a vacuole marker; S14-1, a ribosome marker; or H1, a nucleus marker. For detection of PMA2, COXII and V-ATPase, 5 ?g protein was loaded per lane; for detection of S14-1 and H1, 10 ?g protein was loaded per lane. b Summary of proteins with transmembrane domain (TMD) predicted with use of HMMTOP 2.0. c Venn diagram depicting the distribution of proteins with different lipid modifications. GPI, glycosylphosphatidylinositol anchor; Pre, prenylation site; Myr, myristoylation site; Pal, palmitoylation site. d Proteins predicted to have a TMD or post-translational modification (PTM) or both. e Protein subcellular locations annotated by Gene Ontology or WoLF PSORT showed that 1,121 of the 1,797 proteins with a TMD and/or PTM had PM location information
Reactant: Cucumis sativus (Cucumber)
Application: Western Blotting
Pudmed ID: 30304441
Journal: J Exp Bot
Figure Number: 2B
Published Date: 2019-01-01
First Author: Migocka, M., Maciaszczyk-Dziubinska, E., et al.
Impact Factor: 6.088
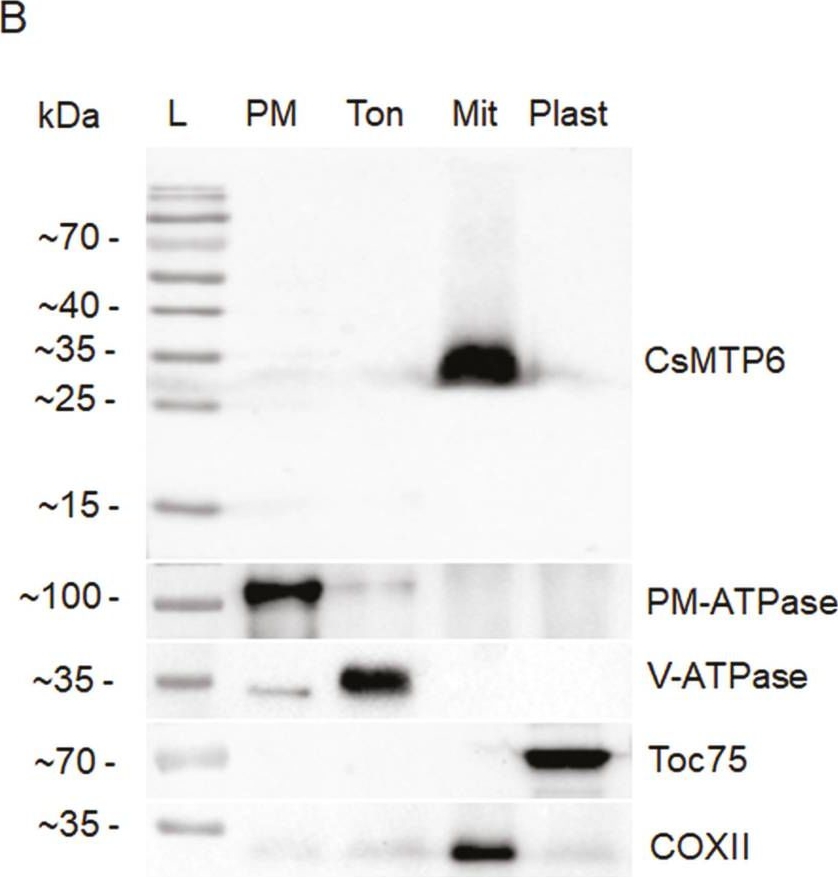
Open PublicationSubcellular localization of CsMTP6. (A) Mitochondrial localization of CsMTP6 in protoplasts prepared from suspensions of Arabidopsis cells: 1, GFP fluorescence of protoplast expressing CsMTP6-GFP; 2, MitoTracker red fluorescence; 3, merged image; 4, transmission image of the same protoplast. The scale bar is 10 µm. (B) Immunolocalization of CsMTP6 in cucumber root cells. SDS-PAGE followed by western blot analysis of plasma membrane (PM), tonoplast (Ton), and mitochondrial (Mit) and plastidial (Plast) fractions. The fractions (15 ?g of protein per lane) were blotted with the primary antibodies to identify marker enzymes for the plasma membrane (PM-ATPase), tonoplast (V-ATPase), plastids (Toc75), mitochondria (COXII), and to localize CsMTP6. L, protein ladder.
Reactant: Cucumis sativus (Cucumber)
Application: Western Blotting
Pudmed ID: 30304441
Journal: J Exp Bot
Figure Number: 7A
Published Date: 2019-01-01
First Author: Migocka, M., Maciaszczyk-Dziubinska, E., et al.
Impact Factor: 6.088
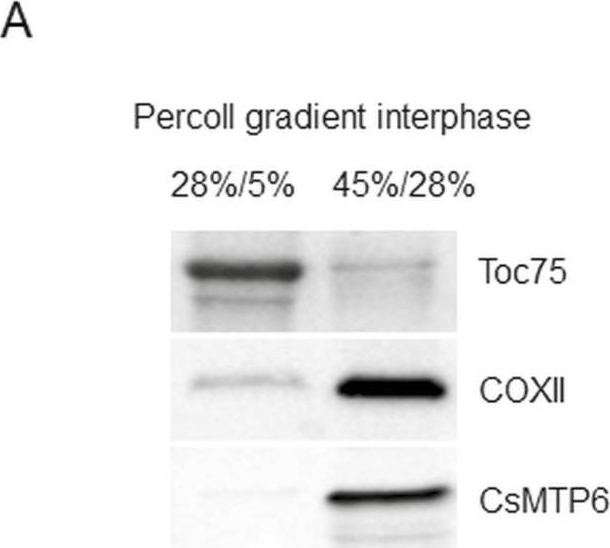
Open PublicationEffects of CsMTP6 expression on mitochondrial Fe and Mn contents and Mn-SOD activity in Arabidopsis protoplasts. (A) Western blot analysis of the purity of mitochondrial and plastidial fractions prepared from protoplasts expressing CsMTP6 using primary antibodies raised against GFP (to detect CsMTP6-GFP), and marker proteins for plastids (Toc75) and mitochondria. The mitochondria and plastid fractions were prepared from protoplasts by discontinuous density gradient centrifugation in Percoll. (B) Fe and (C) Mn content in the mitochondrial fraction prepared from Arabidopsis protoplasts expressing CsMTP6 or transformed with the empty vector. Data are means (±SD) from three separate experiments. (D) Mitochondrial Mn-SOD activity in protoplasts expressing CsMTP6. Data are means (±SD) from three separate experiments, expressed as enzyme units (U). Significant differences were determined using Student’s t-test.

Reactant: Cucumis sativus (Cucumber)
Application: Western Blotting
Pudmed ID: 30304441
Journal: J Exp Bot
Figure Number: 8B
Published Date: 2019-01-01
First Author: Migocka, M., Maciaszczyk-Dziubinska, E., et al.
Impact Factor: 6.088
Open PublicationThe levels of CsMTP6 transcript and protein under different Fe and Mn availability. (A) Real-time analysis of CsMTP6 expression in the roots of 2-week-old cucumber seedlings growing under control Fe (75 µM), control Mn (0.1 µM), excess Fe (1 mM), excess Mn (10 µM), Fe deficiency, or Mn deficiency. Data are mean CsMTP6 transcript levels (±SD) relative to the constitutively expressed reference gene CACS calculated from the arithmetic means of ?Cp values obtained in three independent experiments. (B) Western blot analysis of CsMTP6 protein levels in mitochondria isolated from roots of cucumbers grown under different Fe and Mn treatments for 2 weeks. Immunoblots of the same mitochondria were probed with antibodies against mitochondrial COXII cytochrome oxidase to ensure equal loading of membrane protein (10 µg) onto SDS-PAGE. Significant differences were determined using Tukey’s test.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 32457324
Journal: Sci Rep
Figure Number: 4C
Published Date: 2020-05-26
First Author: Sako, K., Futamura, Y., et al.
Impact Factor: 4.13
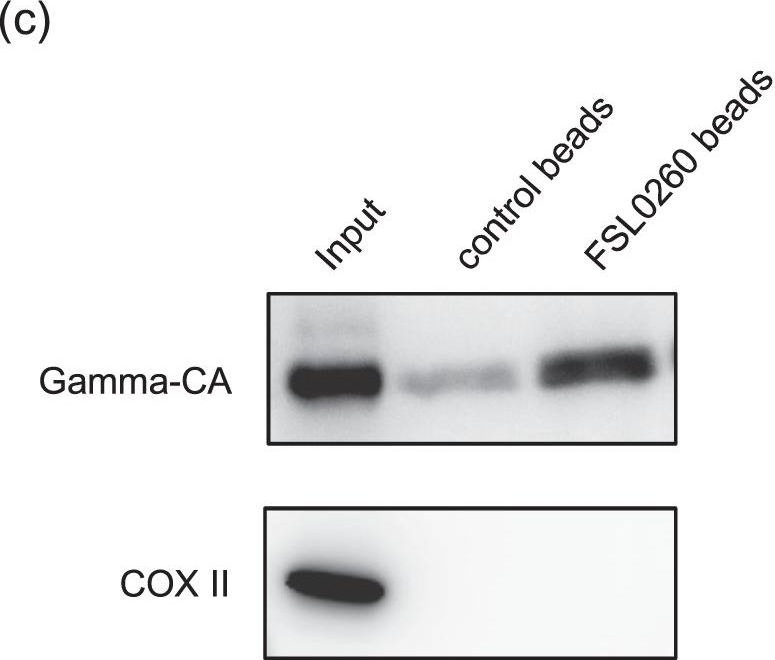
Open PublicationFSL0260 interacts with mitochondrial complex I and inhibits its activity. (a) Inhibition of complex I by FSL0260. Oxygen consumption rate (OCR) of isolated mitochondria from potato tuber was monitored with deamino-NADH in the absence or presence of FSL0260. (b) Inhibition of deamino-NADH oxidation by FSL0260. Deamino-NADH oxidation was measured by spectrometry using sonicated mitochondria isolated from the potato tuber. The experiment was conducted using three biological replicates. Error bars represent the mean ± SE (n?=?3). (c) Pull-down assay of FLS0260. Sonicated potato mitochondria were incubated with control or FSL0260 beads. Immunoblot assay was performed using an anti-gamma CA antibody and an anti-COX II antibody.
- Additional Information
-
Additional information: Cellular [compartment marker] of mitochondrial inner membrane
Additional information (application): Antibody detects COXII protein most optimally in membrane fractions. The signal is weak in a in total protein extract.Blue Native gel electrophoresis (BN-PAGE) has been performed on samples solubilized with digitonin (4:1) and loaded at 100 µg/well. Gel thickness was 2 mm with 4.5-16 % gradient. - Background
-
Background: Cytochrome c oxidase (COX) catalyzes the reduction of oxygen to water in the respiratory chain in the inner mitochondrial membrane. Subunits 1-3 form the functional core of the enzyme complex. Subunit 2 (COXII) transfers the electrons from cytochrome c via its binuclear copper A center to the bimetallic center of the catalytic subunit 1. Alternative name: cytochrome c oxidase subunit 2
- Product Citations
-
Selected references: Shi et al. (2025). Quantitative Analyses of Programmed Cell Death (PCD) Relevant Indicators in Postharvest Vegetable. Future Postharvest and Food. Doi.org/10.1002/fpf2.70027.
Sharma et al. (2025). Comprehensive endoplasmic reticulum proteomics analysis in chickpea (Cicer arietinum L.): unveiling cellular secrets. Journal of Proteins and Proteomics, doi.org/10.1007/s42485-025-00173-z
Ciesielska et al. (2024). S2P2-the chloroplast-located intramembrane protease and its impact on the stoichiometry and functioning of the photosynthetic apparatus of A. thaliana. Front Plant Sci. 2024 Mar 15:15:1372318. doi: 10.3389/fpls.2024.1372318.
Xue et al.(2023). The PtdIns3P phosphatase MtMP promotes symbiotic nitrogen fixation via mitophagy in Medicago truncatula. iScience. 2023 Sep 15;26(10):107752.doi: 10.1016/j.isci.2023.107752.
Leister et al (2023) An ancient metabolite damage-repair system sustains photosynthesis in plants
Suanno et al. (2023) Small extracellular vesicles released from germinated kiwi pollen (pollensomes) present characteristics similar to mammalian exosomes and carry a plant homolog of ALIX, Front. Plant Sci., 25 January 2023, Sec. Plant Membrane Traffic and Transport, Volume 14 - 2023
Inoue et al. (2023) Temperature dependence of O2 respiration in mangrove leaves and roots: implications for seedling dispersal phenology. New Phytol. 2023;237(1):100-112. doi:10.1111/nph.18513
Oikawa et al. (2022) Pexophagy suppresses ROS-induced damage in leaf cells under high-intensity light. Nat Commun. 2022;13(1):7493. Published 2022 Dec 5. doi:10.1038/s41467-022-35138-z
Singh, Muthamilarasan, Prasad (2022). SiHSFA2e regulated expression of SisHSP21.9 maintains chloroplast proteome integrity under high temperature stress. Cell Mol Life Sci. 2022;79(11):580. Published 2022 Nov 3. doi:10.1007/s00018-022-04611-11
Nguyen et al. (2022). MISF2 Encodes an Essential Mitochondrial Splicing Cofactor Required for nad2 mRNA Processing and Embryo Development in Arabidopsis thaliana. Int J Mol Sci. 2022 Feb 28;23(5):2670. doi: 10.3390/ijms23052670. PMID: 35269810; PMCID: PMC8910670.
Gruttner et al. (2022) The P-type pentatricopeptide repeat protein DWEORG1 is a non-previously reported rPPR protein of Arabidopsis mitochondria. Sci Rep. 2022 Jul 21;12(1):12492. doi: 10.1038/s41598-022-16812-0. PMID: 35864185; PMCID: PMC9304396.
Hofmann, Wienkoop & Luthje (2022) Hypoxia-Induced Aquaporins and Regulation of Redox Homeostasis by a Trans-Plasma Membrane Electron Transport System in Maize Roots. Antioxidants (Basel). 2022 Apr 25;11(5):836. doi: 10.3390/antiox11050836. PMID: 35624700; PMCID: PMC9137787.
Kumar et al. (2022). Proteomic dissection of rice cytoskeleton reveals the dominance of microtubule and microfilament proteins, and novel components in the cytoskeleton-bound polysome, Plant Physiology and Biochemistry, Volume 170,2022,Pages 75-86,ISSN 0981-9428, https://doi.org/10.1016/j.plaphy.2021.11.037.
Pavlovic & Kocab. (2021) Alternative oxidase (AOX) in the carnivorous pitcher plants of the genus Nepenthes: what is it good for? Ann Bot. 2021 Dec 18:mcab151. doi: 10.1093/aob/mcab151. Epub ahead of print. PMID: 34922341.
Makino et al. (2020). Induction of Terminal Oxidases of Electron Transport Chain in Broccoli Heads Under Controlled Atmosphere Storage. Foods, 9 (4)
Wang et al. (2020) Rerouting of ribosomal proteins into splicing in plant organelles. BioRxiv, DOI: 10.1101/2020.03.03.974766.
Barua et al. (2019). Dehydration-responsive nuclear proteome landscape of chickpea (Cicer arietinum L.) reveals phosphorylation-mediated regulation of stress response. Plant Cell Environ. 2019 Jan;42(1):230-244. doi: 10.1111/pce.13334.
Waltz et al. (2019). Small is big in Arabidopsis mitochondrial ribosome. Nat Plants. 2019 Jan;5(1):106-117. doi: 10.1038/s41477-018-0339-y.
Shull et al. (2019). Anatase TiO2 nanoparticles induce autophagy and chloroplast degradation in thale cress (Arabidopsis thaliana). Environ Sci Technol. 2019 Jul 29. doi: 10.1021/acs.est.9b01648.
Wang et al. (2019). SMALL KERNEL4 is required for mitochondrial cox1 transcript editing and seed development in maize. J Integr Plant Biol. 2019 Jul 23. doi: 10.1111/jipb.12856.
Chen et al. (2019). PPR-SMR1 is required for the splicing of multiple mitochondrial introns and interacts with Zm-mCSF1 and is essential for seed development in maize. J Exp Bot. 2019 Jun 28. pii: erz305. doi: 10.1093/jxb/erz305.
Waltz et al. (2019). Small is big in Arabidopsis mitochondrial ribosome. Nat Plants. 2019 Jan;5(1):106-117. doi: 10.1038/s41477-018-0339-y.
Gayen et al. (2018). Dehydration-induced proteomic landscape of mitochondria in chickpea reveals large-scale coordination of key biological processes. J Proteomics. 2018 Sep 19. pii: S1874-3919(18)30349-X. doi: 10.1016/j.jprot.2018.09.008
Barua et al. (2018). Dehydration-responsive nuclear proteome landscape of chickpea (Cicer arietinum L.) reveals phosphorylation-mediated regulation of stress response. Plant Cell Environ. 2018 May 10. doi: 10.1111/pce.13334.
Migocka et al. (2018). Cucumber metal tolerance protein 7 (CsMTP7) is involved in the accumulation of Fe in mitochondria under Fe excess. Plant J. 2018 Jun 22. doi: 10.1111/tpj.14006.
Dai et al. (2018). Maize Dek37 Encodes a P-Type PPR Protein That Affects Cis-splicing of Mitochondrial nad2 Intron 1 and Seed Development. Genetics. 2018 Jan 4. pii: genetics.300602.2017. doi: 10.1534/genetics.117.300602.
Nagel et al. (2017). Arabidopsis SH3P2 is an ubiquitin-binding protein that functions together with ESCRT-I and the deubiquitylating enzyme AMSH3. Proc Natl Acad Sci U S A. 2017 Aug 7. pii: 201710866. doi: 10.1073/pnas.1710866114.
Garmash et al. (2017). Expression profiles of genes for mitochondrial respiratory energy-dissipating systems and antioxidant enzymes in wheat leaves during de-etiolation. J Plant Physiol. 2017 Aug;215:110-121. doi: 10.1016/j.jplph.2017.05.023.
Weissenberger et al. (2017). The PPR protein SLOW GROWTH 4 is involved in editing of nad4 and affects the splicing of nad2 intron 1. Plant Mol Biol. 2017 Mar;93(4-5):355-368. doi: 10.1007/s11103-016-0566-4.
Cai et al. (2017). Emp10 encodes a mitochondrial PPR protein that affects the cis-splicing of nad2 intron 1 and seed development in maize. Plant J. 2017 Mar 27. doi: 10.1111/tpj.13551.
Schimmeyer et al. (2016). L-Galactono-1,4-lactone dehydrogenase is an assembly factor of the membrane arm of mitochondrial complex I in Arabidopsis. Plant Mol Biol. 2016 Jan;90(1-2):117-26. doi: 10.1007/s11103-015-0400-4. Epub 2015 Oct 31.
Li et al. (2016). Characterization of a novel Beta-barrel protein (AtOM47) from the mitochondrial outer membrane of Arabidopsis thaliana. J Exp Bot. 2016 Nov;67(21):6061-6075. Epub 2016 Oct 6.
Pavlovic et al. (2016). Light-induced gradual activation of photosystem II in dark-grown Norway spruce seedlings. Biochim Biophys Acta. 2016 Feb 18. pii: S0005-2728(16)30028-7. doi: 10.1016/j.bbabio.2016.02.009.
Li et al. (2015). Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell. 2015 May;27(5):1389-408. doi: 10.1105/tpc.15.00158. Epub 2015 May 5. - Protocols
-
Preparation of plant mitochondrial fraction
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extracts - Reviews:
-
Jane Lee | 2023-12-07This antibody works very well for our samples.G.Vanlerberghe | 2018-02-21This antibody worked effectively in Nicotiana tabacum, using isolated leaf mitochondria.Michal RUREK | 2014-02-1410 µg of mitochondrial proteins from cauliflower (Brassica oleracea var. botrytis) curds (apical layer) was separated on 12 % SDS-PAGE (Laemmli- type) and blotted 1h to Immobilone P (Millipore) using Sedryt apparatus (Kucharczyk). After blocking (5% milk in PBS-T) for 1 h at RT, blots were incubated in the primary antibody at a dilution of 1: 1000 for overnight at 4 deg. with agitation. The primary antibody, diluted in 2% milk in PBS-T was reused several times. Secondary, anti-rabbit HRP- linked antibodies were bound at 1/10000 dilution at RT (1h). Single band of ca. 30 kDa was detected using standart GE Healthcare ECL reagents.Gerhard Obermeyer | 2009-11-18The antibody worked well in Western blots and gave a main signal at ca. 30 kDa in a microsomal fraction prepared from Lilium pollen grains. Anti-COXII was diluted 1:1,000 and an anti-chicken IgY HRP-conjugated (1:10,000) was used for detection.David Macherel | 2009-07-23The antibody yields a clean signal on western blots with extracts from germinating Arabidopsis seeds, young seedlings and leaves. We use it at 1/1000 dilution followed by ECL detection