1

Anti-Enolase 2
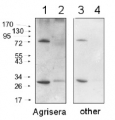
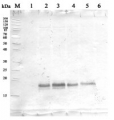
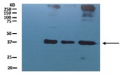
AS10 651 | Clonality:Polyclonal | Host:Rabbit | Reactivity: Arabidopsis thaliana, Helianthus annuus
- Product Info
-
Immunogen: Recombinant Arabidopsis thaliana enolase UniProt: P25696-1, TAIR: At2g36530
Host: Rabbit Clonality: Polyclonal Purity: Serum Format: Lyophilized Quantity: 200 µl Reconstitution: For reconstitution add 200 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Western blot (WB) Recommended dilution: 1 : 2000 (WB) Expected | apparent MW: 47.7 kDa (Arabidopsis thaliana)
- Reactivity
-
Confirmed reactivity: Arabidopsis thaliana, Helianthus annuus Predicted reactivity: Brassica sp., Chlamydomonas reinhardii, Lycopersicum esculentum, Gossypium mexicanum, Nannochloropsis gaditana, Nicotiana tabacum, Oryza sativa, Physcomitrium patens, Populus balsamifera, Ricinus communis, Zea mays
Species of your interest not listed? Contact usNot reactive in: No confirmed exceptions from predicted reactivity are currently known - Application Examples
-
Application example 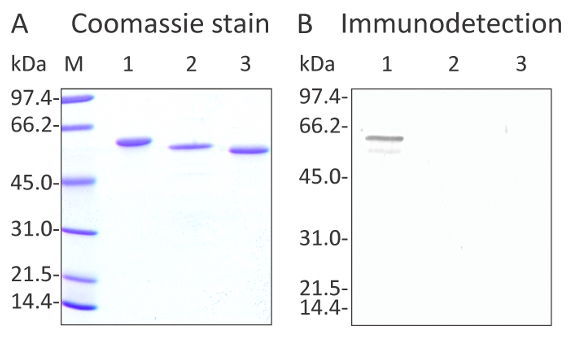
Coomassie staining of three recombinant sunflower ENO proteins after purification on IMAC column and SDS PAGE separation (A) Immunodetection carried out with the anti-Enolase antibody (B) (AS10 651 at 1:2000 dilution). The detection was done with the Goat Anti-Rabbit IgG (H+L) Alkaline phosphatase conjugated (AS09 607 at 1:5000 dilution). In (A) and (B), the lanes were loaded as follows: M indicates the molecular weight markers Lane 1- Recombinant (6xHis)HaENO2 (cytosolic and active isoform) Lane 2- Recombinant (6xHis)ΔHaENO1 (plastidial isoform with the N-terminal transit peptide removed) Lane 3- Recombinant (6xHis)HaENO3 (cytosolic and inactive isoform) In panel (A), 0.7 µg protein was loaded per lane. In panel (B) 50 ng protein was loaded per lane. The faint band seen below the main band in lane 1 in (B) is likely a degradation product of the recombinant protein. No band was detected in lanes 2 and 3.
Recombinant sunflower enolases are described Troncoso-Ponce et al. Plant Science (2018) 272:117-130).
Courtesy of Dr. Jean Rivoal, IRBV, Université de Montréal, Canada - Additional Information
-
Additional information (application): Antibody is specific for the ENO2 isoform (cytosolic and active isoform), see data below - Background
-
Background: Enolase (2-phospho-D-glycerate hydrolase or phosphopyruvate dehydratase) is an essential glycolytic metalloenzyme. It is catalyzing the interconversion of 2-phosphoglycerate and phosphoenolpyruvate.
Alternative name: Bifunctional enolase 2/transcriptional activator - Product Citations
-
Selected references: Zhang et al. (2020). A moonlighting role for enzymes of glycolysis in the co-localization of mitochondria and chloroplasts. Nat Commun. 2020 Sep 9;11(1):4509.doi: 10.1038/s41467-020-18234-w.
Zhang et al. (2018). Nitric oxide induces monosaccharide accumulation through enzyme S-nitrosylation. Plant Cell Environ. 2017 Sep;40(9):1834-1848. doi: 10.1111/pce.12989.
Chen et al.(2009) System analysis of an Arabidopsis mutant altered in de novo fatty acid synthesis reveals diverse changes in seed composition and metabolic regulation. Plant Physiol.
- Protocols
-
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extracts
Oxygenic photosynthesis poster by prof. Govindjee and Dr. Shevela
Z-scheme of photosynthetic electron transport by prof. Govindjee and Dr. Björn and Dr. Shevela - Reviews:
-
This product doesn't have any reviews.