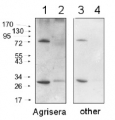
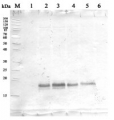
1
Anti-PsbO | 33 kDa of the oxygen evolving complex (OEC) of PSII (anti-peptide)
AS05 092 | Clonality: Polyclonal | Host: Rabbit | Reactivity: A. thaliana, H. vulgare, N. tabacum, P. sativum, Z. mays
- Product Info
-
Immunogen: N-terminually located peptide chosen from Arabidopsis thaliana PsbO1 and PsbO2 isoforms. UniProt: P23321 , PsbO1, TAIR: At5g66570 ; UniProt:A0A178VBH5, TAIR: At3g50820 Host: Rabbit Clonality: Polyclonal Purity: Serum Format: Lyophilized Quantity: 50 µl Reconstitution: For reconstitution add 100 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Immunohistochemistry (IHC), Immunoprecipitation (IP), Western blot (WB) Recommended dilution: 1 : 1000 (WB) Expected | apparent MW: 33 kDa
- Reactivity
-
Confirmed reactivity: Arabidopsis thaliana, Cucumis sativus, Hordeum vulgare, Manihot esculenta, Nicotiana tabacum, Pisum sativum, Sinapsis alba, Triticum aestivum, Zea mays Predicted reactivity: Brassica oleracea, Pisum sativum, Populus tremula, Picea sitchensis, Solanum tuberosum, Vitis vinifera
Species of your interest not listed? Contact usNot reactive in: Chlamydomonas reinhardtii, Synechococcus sp. PCC 7942
For detection of cyanobaterial PsbO, antibody AS21 4689 is suitable. - Application Examples
-
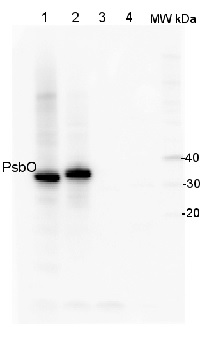
Application example
2 µg of total protein from (1) Arabidopsis thaliana leaf, (2) Horderum vulgare leaf ), (3) Chlamydomonas reinhardtii total cell , (4) Synechococcus sp. 7942 total cell were all extracted with PEB (AS08 300) and separated on 4-12% NuPage (Invitrogen) LDS-PAGE and blotted 1h to PVDF. Blots were blocked immediately following transfer in 2% blocking reagent in 20 mM Tris, 137 mM sodium chloride pH 7.6 with 0.1% (v/v) Tween-20 (TBS-T) for 1h at room temperature with agitation. Blots were incubated in the primary antibody at a dilution of 1: 10 000 for 1h at room temperature with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, then washed once for 15 min and 3 times for 5 min in TBS-T at room temperature with agitation. Blots were incubated in secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated) diluted to 1:50 000 in 2% blocking solution for 1h at room temperature with agitation. The blots were washed as above and developed for 5 min with chemiluminescence detection reagent according to the manufacturers instructions. Images of the blots were obtained using a CCD imager (FluorSMax, Bio-Rad) and Quantity One software (Bio-Rad).
Application examples: 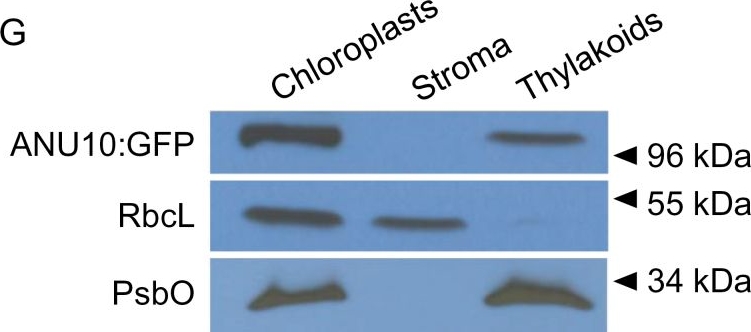
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 24663344
Journal: J Exp Bot
Figure Number: 5G
Published Date: 2014-06-01
First Author: Casanova-Sáez, R., Mateo-Bonmatí, E., et al.
Impact Factor: 6.088
Open PublicationSubcellular and suborganellar localization of ANU10. (A–F) Confocal micrographs of the subepidermal layer of palisade mesophyll cells from (A–C) Ler and (D–F) anu10-1 35Spro:ANU10:GFP transgenic plants. Micrographs show (A, D) the chlorophyll autofluorescence of the chloroplasts, (B, E) the GFP fluorescence, and (C, F) an overlay of the chlorophyll and GFP signals, showing their co-localization in (F). Pictures were taken from first-node leaves collected 16 das. Scale bars indicate 50 ?m. (G) Western blot analysis of the proteins in chloroplast, stroma, and thylakoid fractions isolated from anu10-1 35Spro:ANU10:GFP transgenic plants collected 16 das. Primary antibodies against GFP, the large Rubisco subunit (RbcL), and the PsbO subunit of PSII were used. Molecular mass markers are indicated on the right.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 25835989
Journal: PLoS One
Figure Number: 7A
Published Date: 2015-04-04
First Author: Fristedt, R., Martins, N. F., et al.
Impact Factor: 2.942
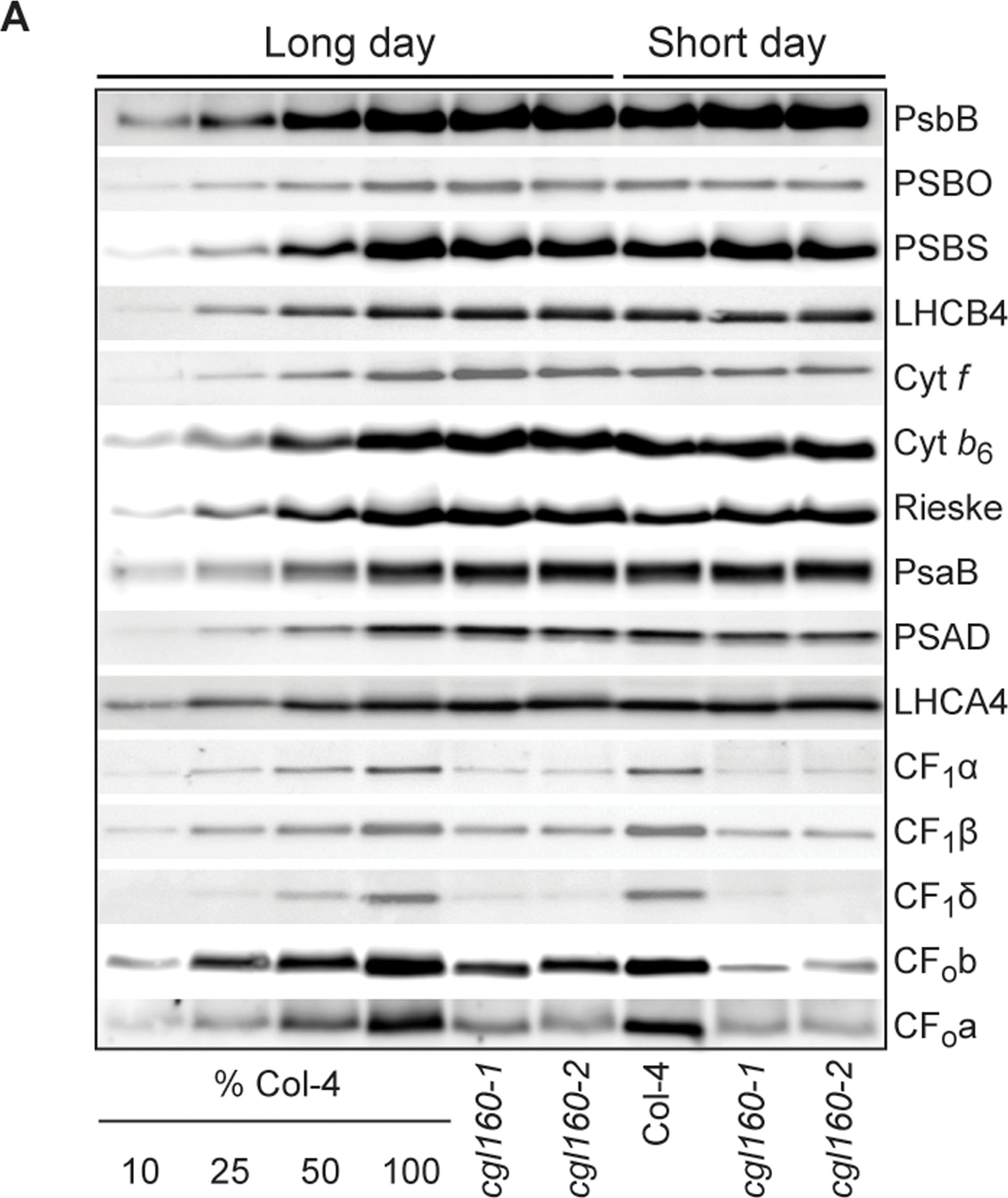
Open PublicationAltered protein accumulation and stability of the chloroplast ATP synthase in the cgl160 mutant visualized by immunoblotting.A. Immunoblots with antibodies against essential subunits of the photosynthetic protein complexes of wild-type (Col-4) Arabidopsis and the two cgl160 T-DNA insertion lines grown under long-day and short-day conditions. Isolated thylakoid membranes were used, and equal amounts of chlorophyll were loaded onto the SDS-PAGE gel. For approximate quantification, wild-type samples from long-day plants were diluted to 10%, 25% and 50%, respectively. Accumulation of PSII was probed with antibodies against PsbB and PSBO. Additionally, the PSBS protein involved in NPQ and the minor PSII antenna protein LHCB4 were probed. Accumulation of the cytochrome b6f complex was probed with antibodies against the essential subunits PetA (cytochrome f), PetB (cytochrome b6), and PETC (Rieske protein). Accumulation of PSI was probed with antibodies against the reaction center subunit PsaB and the stromal ridge subunit PsaD. ATP synthase accumulation was probed with antibodies against the CF1 subunits AtpA (CF1?), AtpB (CF1?) and AtpD (CF1?) and antibodies against the CF0 subunits AtpF (CF0b) and AtpI (CF0a). B. Loading difference estimation for immunoblotting CF1 between wild type and cgl160-1. To obtain similar immunoblotting signal three times more (15 ?g protein) was needed for cgl160-1 compared to wild type (5 ?g protein). C. Maintenance of CF1 was measured by incubating leaves from wild type and cgl160-1 in solution containing the plastid protein synthesis inhibitor chloramphenicol for the indicated time points. Protein extract was isolated and separated by SDS-PAGE, immunoblotted and probed with specific antibodies against CF1 and LHCB2.1. Three times more protein was loaded from the mutant to obtain equal level of CF1 immunoblotting signal, as specified in B.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 26442058
Journal: Front Plant Sci
Figure Number: 6B
Published Date: 2015-10-07
First Author: Murgia, I., Giacometti, S., et al.
Impact Factor: 5.435
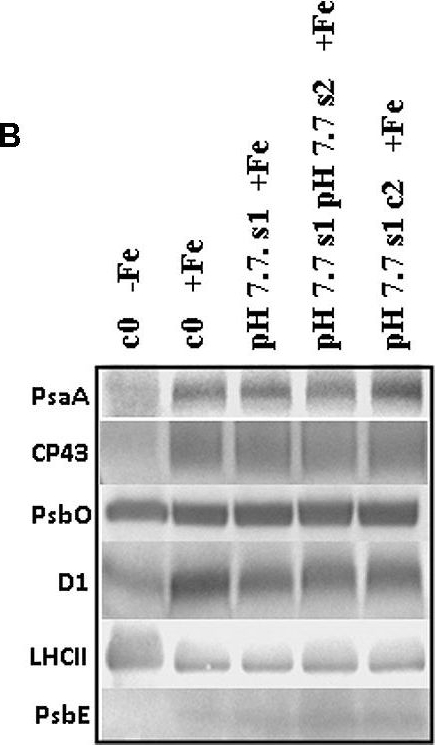
Open PublicationO2 evolution and protein profile of photosynthetic apparatus in A. thaliana seedlings with generational exposure to Fe deficiency. (A) SHR-trap 1445 c0, pH 7.7 s1 pH 7.7 s2, pH 7.7 s1 c2 seedlings were grown for 11 days in control AIS medium and net O2 evolution (expressed as ?mol O2 evolved min-1 mg chlorophyll-1) was measured under illumination at either 100 or 800 ?E m-2 s-1. Values are mean ± SE of three biological replicas, each consisting of at least 15 seedlings. Letters represent statistical differences, according to Student’s t-test, with p < 0.05. (B) Western blot analysis with antibodies against PsaA, CP43, PsbO, D1, LHCII, and PsbE of thylakoid membranes purified from 11 days-old SHR-trap 1445 c0 seedlings germinated in -Fe AIS medium or from 11 days-old SHR-trap 1445 c0, pH 7.7 s1, pH 7.7 s1 pH 7.7 s2, pH 7.7 s1 c2 seedlings germinated in control (+Fe) AIS medium. Samples corresponding to 1 ?g chlorophyll were loaded on each lane.
Reactant: Plant
Application: Western Blotting
Pudmed ID: 27590049
Journal: BMC Plant Biol
Figure Number: 9A
Published Date: 2016-09-02
First Author: Mazur, R., Sadowska, M., et al.
Impact Factor: 4.142
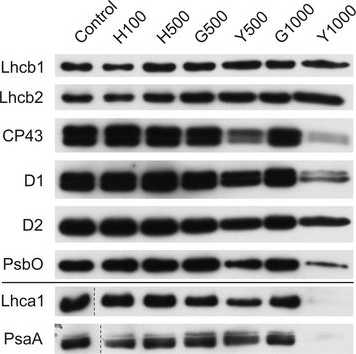
Open PublicationChanges of PSII and PSI antenna and core protein levels. Proteins from control and Tl-treated white mustard leaves were separated by SDS-PAGE followed by immunodetection with antibodies against Lhcb1, Lhcb2, Lhca1 (antenna proteins) and D1, D2, CP43, PsbO, PsaA (core proteins). Samples were loaded on the equal amount of chlorophyll (0.25 ?g). Description of samples abbreviation as given in the legend to Fig. 3
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 28791032
Journal: Front Plant Sci
Figure Number: 2A
Published Date: 2017-08-10
First Author: Kohzuma, K., Froehlich, J. E., et al.
Impact Factor: 5.435
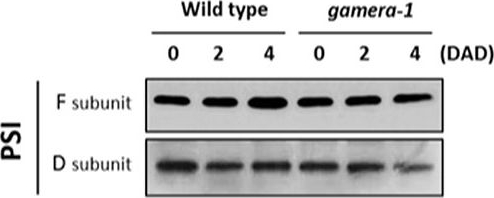
Open PublicationChanges in the protein levels of photosynthetic components under extended dark exposure in wild-type and gamera-1. Immunoblot detection of photosynthetic proteins from leaves of Ws and gamera-1 plants incubated after dark adaptation for 0, 2, and 4 days was examined. Specifically, essentially thylakoid fractions were assayed to determine the content of the following proteins: ?-subunit of ATP synthase; the D1 protein, OEC17, OEC23, and OEC33 of PSII; Cyt f and Rieske protein of the cytochrome b6f complex; and the F and D subunits of PSI, after extended dark treatment. Proteins were resolved via SDS-PAGE gel based on equal microgram chlorophyll per lane loading and processed as described in Section “Materials and Methods”. The Large subunit of RuBisco and LHCII stained with either CBB or Ponceau red, respectively, are presented here as loading controls. DAD indicates days after dark adaptation.
- Additional Information
-
Additional information: Loading based on 50-100 ng of chlorophyll is enough to obtain good signal with this antibody
Additional information (application): Good signal is obtained with this antibody with a load from 0,5 chlorophyll µg/well - Background
-
Background: The PsbO protein is an extrinisic subunit of the water splitting photosystem II (PSII) complex. The protein is exposed on the luminal side of the thylakoid membrane, and is hihgly conserved in all known oxygenic photosynthetic organisms. Alternative names of PsbO1 include 33 kDa subunit of oxygen evolving system of photosystem II, OEC 33 kDa subunit, 33 kDa thylakoid membrane protein, manganese-stabilizing protein 1 and for PsbO2 33 kDa subunit of oxygen evolving system of photosystem II, OEC 33 kDa subunit, 33 kDa thylakoid membrane protein, manganese-stabilizing protein 2.
- Product Citations
-
Selected references: Silvestre et al. (2025). A Holistic Investigation of Arabidopsis Proteomes Altered in Chloroplast Biogenesis and Retrograde Signalling Identifies PsbO as a Key Regulator of Chloroplast Quality Control. Plant Cell Environ. 2025 Aug;48(8):6373-6396. doi: 10.1111/pce.15611.
Zhao et al. (2024). Psb28 protein is indispensable for stable accumulation of PSII core complexes in Arabidopsis.Plant J. 2024 May 26. doi: 10.1111/tpj.16844.
Jin et al. (2023) Dual roles for CND1 in maintenance of nuclear and chloroplast genome stability in plants. Cell Rep. 2023 Mar 28;42(3):112268. doi: 10.1016/j.celrep.2023.112268. Epub 2023 Mar 17.
Xiong et al. (2022) a chloroplast nucleoid protein of bacterial origin linking chloroplast transcriptional and translational machineries, is required for proper chloroplast gene expression in Arabidopsis thaliana. Nucleic Acids Res. 2022 Jun 23;50(12):6715–34. doi: 10.1093/nar/gkac501. Epub ahead of print. PMID: 35736138; PMCID: PMC9262611.
Mazur et al. (2021) The SnRK2.10 kinase mitigates the adverse effects of salinity by protecting photosynthetic machinery. Plant Physiol. 2021 Dec 4;187(4):2785-2802. doi: 10.1093/plphys/kiab438. PMID: 34632500; PMCID: PMC8644180.
Toubiana et al. (2020). Correlation-based Network Analysis Combined With Machine Learning Techniques Highlight the Role of the GABA Shunt in Brachypodium Sylvaticum Freezing Tolerance. Sci Rep , 10 (1), 4489
Wang et al. (2019). YR36/WKS1-mediated Phosphorylation of PsbO, an Extrinsic Member of Photosystem II, Inhibits Photosynthesis and Confers Stripe Rust Resistance in Wheat. Mol Plant. 2019 Oct 14. pii: S1674-2052(19)30330-2. doi: 10.1016/j.molp.2019.10.005.
An et al. (2019). Protein cross-interactions for efficient photosynthesis in the cassava cultivar SC205 relative to its wild species. J Agric Food Chem. 2019 Jul 19. doi: 10.1021/acs.jafc.9b00046.
Rozp?dek et al. (2018). Acclimation of the photosynthetic apparatus and alterations in sugar metabolism in response to inoculation with endophytic fungi. Plant Cell Environ. 2018 Dec 5. doi: 10.1111/pce.13485.
Li et al. (2018). Comparative proteomic analysis of key proteins during abscisic acid-hydrogen peroxide-induced adventitious rooting in cucumber (Cucumis sativus L.) under drought stress. Journal of Plant Physiology Volume 229, October 2018, Pages 185-194.
Mazur et al. (2016). Overlapping toxic effect of long term thallium exposure on white mustard (Sinapis alba L.) photosynthetic activity. BMC Plant Biol. 2016 Sep 2;16(1):191. doi: 10.1186/s12870-016-0883-4.
Albanese et al.(2016). Isolation of novel PSII-LHCII megacomplexes from pea plants characterized by a combination of proteomics and electron microscopy. Photosynth Res. 2016 Jan 9.
Fristedt et al. (2015). The thylakoid membrane protein CGL160 supports CF1CF0 ATP synthase accumulation in Arabidopsis thaliana. PLoS One. 2015 Apr 2;10(4):e0121658. doi: 10.1371/journal.pone.0121658.
Hu et al. (2015). Site-specific Nitrosoproteomic Identification of Endogenously S-Nitrosylated Proteins in Arabidopsis. Plant Physiol. 2015 Feb 19. pii: pp.00026.2015.
Casanova-Saez et al. (2014). Arabidopsis ANGULATA10 is required for thylakoid biogenesis and mesophyll development. J Exp Bot. 2014 Mar 24.
Albus et al. (2012). LCAA, a novel factor required for Mg protoporphyrin monomethylester cyclase accumulation and feedback-control of aminolevulinic acid biosynthesis in tobacco. Plant Physiol. Oct 19. - Protocols
-
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extracts - Reviews:
-
Wioleta Wasilewska | 2014-12-17I used this antibody for 33 kDa protein immunodetection analysis in three types of chloroplasts from two plant species: Zea mays and Pisum sativum. Equivalent of 1.0 µg of thylakoids extract were loaded on SDS- PAGE (12% acrylamide). In all types of chloroplasts I observed high quality signal and I had no problems with Agrisera antibody. The product was efficient and I recommend it for people working with photosystem II.Jerzy Kruk | 2013-11-21Analysis perfomed on Arabidopsis total leaf extract, 10% SDS gel, 3 ug chlorophyll per lane, AB dilution 1:2000, the product observed at ca. 33 kDaYu Qing-Bo | 2009-05-07This antibody works very well in our lab, when it was used to detect the accumulation of photosynthetic protein in Arabidopsis