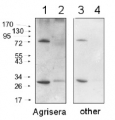
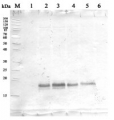
1
Anti-RFA | Baker's yeast replication factor A
AS07 214 | Clonality: Polyclonal | Host: Rabbit | Reactivity: Saccharomyces cerevisiae
- Product Info
-
Immunogen: RPA from Saccharomyces cerevisiae consisting of three subunits RFA1 (70 kDa), RFA2 (30 kDa) and RFA3 (14 kDa); overexpressed in E.coli and purified by chromatography; no affinity tags were added to any of three subunits
Host: Rabbit Clonality: Polyclonal Purity: Serum Format: Lyophilized Quantity: 50 µl Reconstitution: For reconstitution add 50 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Immunoprecipitation (IP), Chromatin Immunoprecipitation (ChIP), Western blot (WB) Recommended dilution: ChIP, 1 : 20 000 (WB) Expected | apparent MW: 70 + 30 + 14 kDa
- Reactivity
-
Confirmed reactivity: Saccharomyces cerevisiae Predicted reactivity: Saccharomyces cerevisiae
Not reactive in: No confirmed exceptions from predicted reactivity are currently known - Application Examples
-
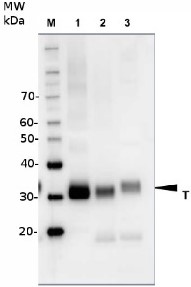
TCA precipitated protein extracts from a wild type yeast strain (S. cerevisiae) were separated on 10% gel and transferred to a PVDF membrane. Antibody was used in different dilutions: 1: 5000 (1); 1: 10 000 (2); 1: 20 000 (3);
Besides the bands for RFA1 and RFA2 an unspecific band was detected at ~150 kDa.
Application examples: 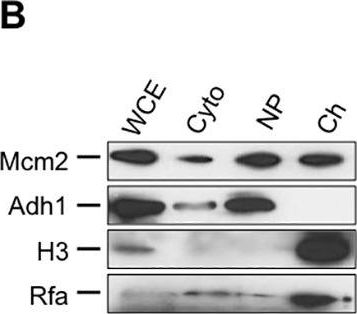 Shi et al. (2023).A R-loop sensing pathway mediates the relocation of transcribed genes to nuclear pore complexes.STAR Protoc. 2023 Sep 20;4(4):102577.doi: 10.1016/j.xpro.2023.102577.
Shi et al. (2023).A R-loop sensing pathway mediates the relocation of transcribed genes to nuclear pore complexes.STAR Protoc. 2023 Sep 20;4(4):102577.doi: 10.1016/j.xpro.2023.102577.Reactant: Saccharomyces cerevisiae (Yeast)
Application: Western Blotting
Pudmed ID: 27923055
Journal: PLoS Genet
Figure Number: 1B
Published Date: 2016-12-01
First Author: Syed, S., Desler, C., et al.
Impact Factor: 5.334
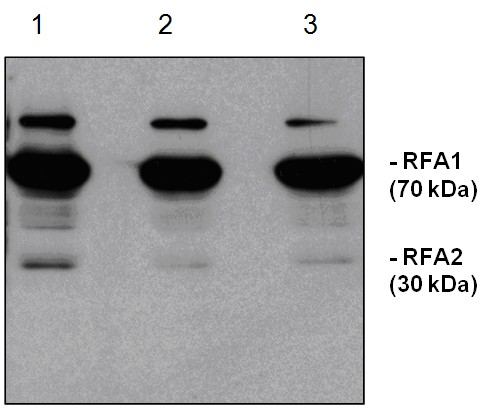
Open PublicationSILAC-based quantification of changes in chromatin association in cells lacking Rrm3.(A) Equal numbers of light-labeled (wildtype) and heavy-labeled (rrm3?) cells were mixed, nuclei isolated, and the chromatin fraction extracted and analyzed by tandem mass spectrometry using a hybrid linear ion trap-Orbitrap instrument. (B) Subcellular fractionation was verified by following the distribution of proteins in cytoplasmic (Cyto), nucleoplasm (NP), and chromatin (Ch) fractions during the enrichment procedure by Western blotting. Successful chromatin fractionation is indicated by the enrichment of histone H3 and Rfa and the absence of Adh1. Chromatin fractions were analyzed from three biological replicates in (C) the absence of hydroxyurea and (D) in the presence of hydroxyurea. (E) Merger of peptide quantification in the absence and presence of hydroxyurea.
- Additional Information
-
Additional information (application): Antibody was also successfully used in ChIP application Holstein et al. (2014).
Load of 1 ng of the protein will allow to visualize two subunits of RPA, while load of 5 ng will allow to visualize all three subunits in Western blot technique. - Background
-
Background: Saccharomyces cerevisiaereplication protein A (RPA), also known as replication factor A (RFA) is a single-stranded DNA-binding protein that is required for multiple processes in eukaryotic DNA metabolism. Those processes include DNA replication, DNA repair, and recombination. Homologues to RPA have been identified in all eukaryotic organisms examined. RPA is heterotrimeric protein composed of subunits of approximately 70, 30, and 14 kDa. Members of this family bind nonspecifically to single-stranded DNA and interact with and/or modify the activities of multiple proteins. Alternative names: Replication protein A 69 kDa DNA-binding subunit, Single-stranded DNA-binding protein, DNA-binding protein BUF2, replication protein A 36 kDa subunit, DNA-binding protein BUF1 antibody
- Product Citations
-
Selected references: Choi et al. (2025). Recombination-coupled DNA synthesis facilitates post-invasion steps in meiotic crossover and noncrossover formations. Nucleic Acids Res. 2025 Jul 19;53(14):gkaf704. doi: 10.1093/nar/gkaf704.
Masnovo et al. (2024). Stabilization of expandable DNA repeats by the replication factor Mcm10 promotes cell viability. Nat Commun . 2024 Dec 3;15(1):10532. doi: 10.1038/s41467-024-54977-6.
Galanti et al. (2024). Dbf4-dependent kinase promotes cell cycle controlled resection of DNA double-strand breaks and repair by homologous recombination. Nat Commun. 2024 Apr 3;15(1):2890. doi: 10.1038/s41467-024-46951-z.
Kotenko and Makovets (2024).The functional significance of the RPA- and PCNA-dependent recruitment of Pif1 to DNA. EMBO Rep. 2024 Mar 13. doi: 10.1038/s44319-024-00114-9.
Joo et al. (2024).RPA interacts with Rad52 to promote meiotic crossover and noncrossover recombination. Nucleic Acids Res. 2024 Feb 10:gkae083.doi: 10.1093/nar/gkae083.
Shi et al. (2023).A R-loop sensing pathway mediates the relocation of transcribed genes to nuclear pore complexes.STAR Protoc. 2023 Sep 20;4(4):102577.doi: 10.1016/j.xpro.2023.102577.
Zhai et al (2023) Dna2 removes toxic ssDNA-RPA filaments generated from meiotic recombination-associated DNA synthesis
Pires, Lohner, Wagner, et al. (2023) RNA-DNA hybrids prevent resection at dysfunctional telomeres [published online ahead of print, 2023 Feb 1]. Cell Rep. 2023;42(2):112077. doi:10.1016/j.celrep.2023.112077.
Reusswig, et al. (2022) Unscheduled DNA replication in G1 causes genome instability and damage signatures indicative of replication collisions. Nat Commun. 2022;13(1):7014. Published 2022 Nov 18. doi:10.1038/s41467-022-34379-2.
Scherzer et al (2022). Recruitment of Scc2/4 to double-strand breaks depends on ?H2A and DNA end resection. Life Sci Alliance. 2022 Jan 27;5(5):e202101244. doi: 10.26508/lsa.202101244. PMID: 35086935; PMCID: PMC8807874.
Minchell et al. (2020). Cohesin Causes Replicative DNA Damage by Trapping DNA Topological Stress. Mol Cell . 2020 Mar 29;S1097-2765(20)30161-1. doi: 10.1016/j.molcel.2020.03.013.
He et. al (2019). KEOPS complex promotes homologous recombination via DNA resection. Nucleic Acids Res. 2019 Apr 2. pii: gkz228. doi: 10.1093/nar/gkz228.
Jakobsen et al. (2019). Minimal Resection Takes Place during Break-Induced Replication Repair of Collapsed Replication Forks and Is Controlled by Strand Invasion. Cell Rep. 2019 Jan 22;26(4):836-844.e3. doi: 10.1016/j.celrep.2018.12.108. (used AS07 214-100, which is a larger size unit of AS07 214)
Deshpande et al. (2017). Structural Basis of Mec1-Ddc2-RPA Assembly and Activation on Single-Stranded DNA at Sites of Damage. Mol Cell. 2017 Oct 19;68(2):431-445.e5. doi: 10.1016/j.molcel.2017.09.019.
Chen et al. (2017). Dihydrocoumarin, an HDAC Inhibitor, Increases DNA Damage Sensitivity by Inhibiting Rad52. Int J Mol Sci. 2017 Dec 7;18(12). pii: E2655. doi: 10.3390/ijms18122655.
Yeeles et al. (2015). Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015 Mar 4. doi: 10.1038/nature14285.
Holstein et al. (2014). Interplay between Nonsense-Mediated mRNA Decay and DNA Damage Response Pathways Reveals that Stn1 and Ten1 Are the Key CST Telomere-Cap Components. Cell Rep. 2014 May 22;7(4):1259-69. doi: 10.1016/j.celrep.2014.04.017. Epub 2014 May 15. (ChIP).
Deng et al. (2014). RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat Struct Mol Biol. 2014 Mar 9. doi: 10.1038/nsmb.2786. (western blot).
Bentensen et al. (2013). MRX protects fork integrity at protein–DNA barriers, and its absence causes checkpoint activation dependent on chromatin contex. Nucleic Acids Res. 2013 Mar 1;41(5):3173-89. doi: 10.1093/nar/gkt051. (ChIP). - Protocols
- Antibody protocols
- Reviews:
-
This product doesn't have any reviews.