1

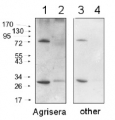
Anti-Rnr3 | Ribonucleoside-diphosphate reductase large chain 2
AS09 574 | Clonality: Polyclonal | Host: Rabbit | Reactivity: Saccharomyces cerevisiae
- Product Info
-
Immunogen: KLH-conjugated synthetic peptide derived from Saccharomyces cerevisiae Rnr3 protein sequence UniProt: P21672
Host: Rabbit Clonality: Polyclonal Purity: Immunogen affinity purified serum in PBS pH 7.4. Format: Lyophilized Quantity: 100 µg Reconstitution: For reconstitution add 100 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Western blot (WB) Recommended dilution: 1 : 500-1 : 1000 (WB) Expected | apparent MW: 97.5 | 98 kDa - Reactivity
-
Confirmed reactivity: Saccharomyces cerevisiae Predicted reactivity: Saccharomyces cerevisiae
Not reactive in: No confirmed exceptions from predicted reactivity are currently known - Application Examples
-
Application example
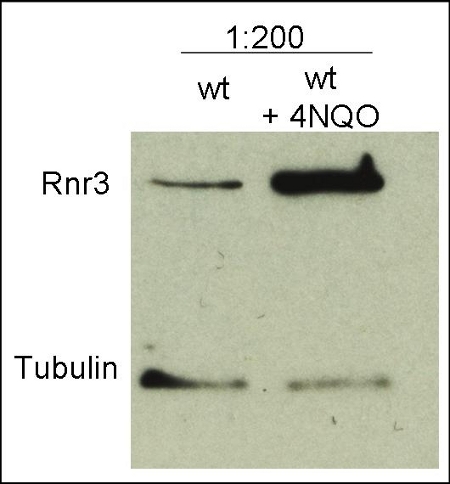
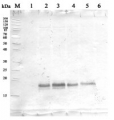
10 μl total protein from 9.25 x 107 cells of Saccharomyces cerevisiae extracted with 20% TCA as described below were separated on 10% SDS-PAGE and blotted 1.5h (0.5 A) to a nitrocellulose membrane (Whatman PROTRAN BA 85, 0.45 μm). Blots were blocked with 5% non-fat dry milk in TBST for 1.5h at room temperature (RT) with agitation. Blot was incubated in the primary antibody at a dilution of 1: 200 overnight at 4C° with agitation. The antibody solution was decanted and the blot was washed 3 times for 10 min in TBST at RT with agitation. Blot was incubated in secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated, (AS09 602) diluted to 1:50 000 for 1h at RT with agitation. The blot was washed as above and developed for 3 min with chemiluminescent detection reagent, according to the manufacturers instructions. Exposure time was 10 min.
Courtesy Dr. Andrei Chabes, Umeå University, Sweden
Application examples: 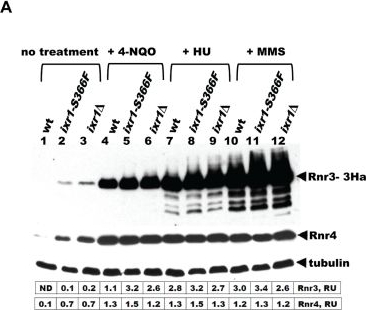
Reactant: Saccharomyces cerevisiae (Yeast)
Application: Western Blotting
Pudmed ID: 21573136
Journal: PLoS Genet
Figure Number: 3A
Published Date: 2011-05-01
First Author: Tsaponina, O., Barsoum, E., et al.
Impact Factor: 5.334
Open PublicationDeletion of IXR1 leads to increased Rnr3 and Rnr4 levels and decreased Sml1 levels.(A) Western blot analysis of Rnr3-HA and Rnr4 levels in wild-type (AC447-2A), ixr1-S366F (TOY619), and ixr1? (TOY621) strains before and after 2 hours treatment with 0.2 mg/L 4-nitroquinoline 1-oxide (4-NQO), 200 mM HU, or 0.02% methyl methanesulfonate (MMS). Rnr3 and Rnr4 levels were quantified in relative units (RU, levels of Rnr3 or Rnr4 divided by the levels of tubulin in corresponding sample) as described in Materials and Methods. ND – not detected. (B) Western blot analysis of Rnr2 levels in wild-type (AC447-2A) and ixr1? (TOY621) strains before treatment and after 2 hours treatment with 0.2 mg/L 4-NQO, 0.02% MMS, or 200 mM HU. (C) Western blot analysis of Sml1 levels in wild-type (W1588-4C) and ixr1? (TOY736) strains before treatment and after 2 hours treatment with 0.02% MMS. (D) Western blot analysis of Rad53 phosphorylation status in wild-type (W1588-4C) and ixr1? (TOY736) strains before treatment and after 2 hours treatment with 0.02% MMS. (E) Deletion of RAD53 but not of SML1 or DUN1 abolishes the upregulation of Rnr3 and Rnr4 levels in ixr1?. Western blot analysis of Rnr3-HA and Rnr4 levels. The following strains were analyzed: wt (AC447-2A), ixr1? (TOY732), ixr1? sml1? (TOY778), ixr1? dun1? sml1? (TOY772), and ixr1? rad53? sml1? (TOY781).
- Additional Information
-
Additional information (application): Load per well was approx 3x10^6 cells (of a total extract) - Background
-
Background: Saccharomyces cerevisiae Rnr3 is catalyzing the biosynthesis of deoxyribonucelaotides. Alternative names: Ribonucleotide reductase large subunit 2, ribonucleotide reductase DNA damage-inducible regulatory subunit 2, ribonucleotide reductase R1 subunit 2
- Product Citations
-
Selected references: Van der Horst et al. (2025). Replication-IDentifier links epigenetic and metabolic pathways to the replication stress response. Nat Commun. 2025 Feb 6;16(1):1416. doi: 10.1038/s41467-025-56561-y.
Ajazi et al. (2021) Endosomal trafficking and DNA damage checkpoint kinases dictate survival to replication stress by regulating amino acid uptake and protein synthesis. Dev Cell. 2021 Sep 27;56(18):2607-2622.e6. doi: 10.1016/j.devcel.2021.08.019. Epub 2021 Sep 16. PMID: 34534458.
Cerritelli et al. (2020). High density of unrepaired genomic ribonucleotides leads to Topoisomerase 1-mediated severe growth defects in absence of ribonucleotide reductase. Nucleic Acids Res
Sampaio-Marques et al. (2019). ?-Synuclein toxicity in yeast and human cells is caused by cell cycle re-entry and autophagy degradation of ribonucleotide reductase 1. Aging Cell. 2019 Aug;18(4):e12922. doi: 10.1111/acel.12922.
Schmidt et al. (2019). Inactivation of folylpolyglutamate synthetase Met7 results in genome instability driven by an increased dUTP/dTTP ratio. Nucleic Acids Res. 2019 Oct 24. pii: gkz1006. doi: 10.1093/nar/gkz1006.
Lafuente-Barquero et al. (2017). The Smc5/6 complex regulates the yeast Mph1 helicase at RNA-DNA hybrid-mediated DNA damage. PLOS Genetics, December 27, 2017, doi.org/10.1371/journal.pgen.1007136
Schmidt et al. (2017). Alterations in cellular metabolism triggered by URA7 or GLN3 inactivation cause imbalanced dNTP pools and increased mutagenesis. Proc Natl Acad Sci U S A. 2017 May 30;114(22):E4442-E4451. doi: 10.1073/pnas.1618714114.
Graf et al. (2017). Telomere Length Determines TERRA and R-Loop Regulation through the Cell Cycle. Cell. 2017 Jun 29;170(1):72-85.e14. doi: 10.1016/j.cell.2017.06.006.
Williams et al. (2017). The role of RNase H2 in processing ribonucleotides incorporated during DNA replication. DNA Repair (Amst). 2017 Mar 6. pii: S1568-7864(16)30431-1. doi: 10.1016/j.dnarep.2017.02.016. - Protocols
-
Recommended western blot protocol for work with Rnr3 antibodies
1. Run 7.5% SDS-PAGE, run out proteins up to 40 kDa in size.
2. Transfer proteins to a nitrocellulose membrane Protran BA 85, Whatman; 1h RT 300V/300 mA.
Transfer buffer: 50 ml 40xNaCarb, 400 ml Ethanol (95,5%), 1450 ml ddH20.
40xNaCarb: 42.19 g NaHCO3, 36.7 g Na2CO3, dissolve in 1.5 L ddH2O, adjust pH to 9.5 with HCl, bring to 1.8 L and autoclave.3. Stain with Ponceau S, cut the membrane between 70 and 85 kDa, and wash away the dye with autoclaved ddH2O. Upper part is used for Rnr3 Western Blot.
4. Block membrane with 5% non-fat milk in 1xTBST, 1.5 h RT
10xTBST 1L: 100 ml 1M Tris pH=8.0, 87.66g NaCl, 5 ml Tween20.
Dilute Ab 1:5000 in 5% non-fat milk in 1x TBST 1:5000, incubate ON +4C (can be stored at -20°C and re-used up to 4 times).5. Wash 3 times in 1xTBST, 10 min each RT
6. Incubate with a secondary Ab 1:5000 in 1xTBST no milk, 1h RT
7. Wash 3 times in 1xTBST, 10 min each RT
8. Develop the membrane.
- Reviews:
-
Duhita Mirikar | 2024-04-24Species- Saccharomyces cerevisiae Application- Western blot Dilution- 1:1000 . Rnr3 antibody is very specific to the protein with no non-specific band. It shows an Intense band