2

Anti-V-ATPase | Epsilon subunit of tonoplast H+ATPase
AS07 213 | Clonality: Polyclonal | Host: Rabbit | Reactivity: Higher plants including A.comosus, A.thaliana, C.sativus, C. australis R.Br, C.reinhardtii, F. margarita Swingle, H.vulgare, L.esculentum, L.longiflorum, Malus x domestica Borkh. c.v. Fuji, M. truncatula, M.crystallinum, N.tabacum, N.caerulescens, O.sativa, P.hybrida cv. Mitchell, Populus sp., P.vittata, Thellungiella sp., T. aestivum, Z.mays, V. vinifera | Cellular [compartment marker] of tonoplast membrane
- Product Info
-
Immunogen: KLH-conjugated synthetic peptide chosen from subunit E of plant V-ATPase including Arabidopsis thaliana UniProt: Q39258-1, TAIR: At4g11150. Peptide is conserved in vacuolar H+-ATPase subunit E, isoform 1 to 3 (VHA-E1). Host: Rabbit Clonality: Polyclonal Purity: Antigen affinity purified serum, in PBS pH 7.4 Format: Lyophilized Quantity: 50 µg Reconstitution: For reconstitution add 50 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Immunofluorescence (IF), Immunohistochemistry (IHC), Western blot (WB) Recommended dilution: 1: 100 (IF), 1 : 50 (IHC), 1 : 2000-1 : 5000 (WB) Expected | apparent MW: 26 | 31 kDa (Arabidopsis thaliana)
- Reactivity
-
Confirmed reactivity: Ananas comosus, Arabidopsis thaliana, Cucumis sativus, Chara australis , Chlamydomonas reinhardtii, Fortunella margarita Swingle, Hordeum vulgare, Lilium longiflorum, Malus x domestica Borkh. c.v. Fuji, Medicago truncatula, Mesembryanthemum crystallinum, Nicotiana tabacum, Noccaea caerulescens, Oryza sativa, Petunia hybrida cv. Mitchell, Populus sp., Pteris vittata (fern), Salicornia bigelovii, Solanum lycopersicum, Thellungiella sp., Triticum aestivum, Zea mays, Vitis vinifera Predicted reactivity: Brachypodium dystachyon, Capsella rubella, Chenopodium quinoa, Citrus clementina, Citrus unshiu, Citrus limon, Eucalypsus grandis, Glyxine max, Glycine soja, Lotus japonicus, Phaseolus sp., Physcomitrium patens, Populus trichocarpa, Prunus persica, Ricinus communis, Riticum aestivum, Solanum tuberosum, Sorghum bicolor Theobroma cacao, Vitis vinifera,
Bull frog, Chicken, Bovine, Drosophila melanogaster, Human, Mouse, Rat
Species of your interest not listed? Contact usNot reactive in: Avicennia sp., mangrove plants, Schizosaccharomyces pombe - Application Examples
-
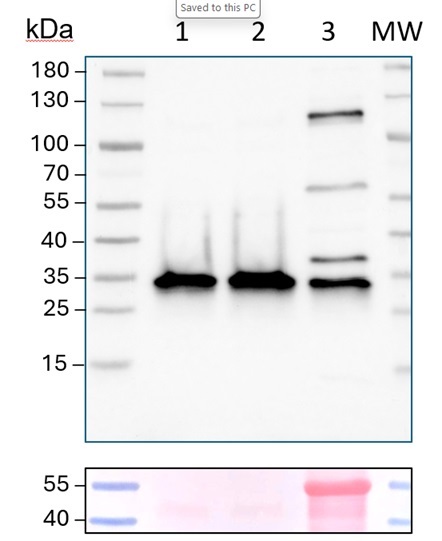
Samples:
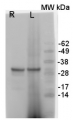
1 - 5 ug of Arabidopsis thaliana wild type, purified vacuoles, replicate 1
2 - 5 ug of Arabidopsis thaliana wild type, purified vacuoles, replicate 2
3 - 20 ug of Arabidopsis thaliana whole leaf extract wild type
5 ug/well for vacuole samples* and 15 µg/well of total cell lysate protein extracted freshly from Arabidopsis thaliana rosette leaves. For vacuole isolation I followed the published protocol https://doi.org/10.1038/nprot.2007.26). Extract buffer components were: 150 mM NaCl, 50 mM Tris-HCl pH 7.5, 0.1% NP-40, 1% Protease inhibitor cocktail, 1% DPDS; and denatured with loading buffer (5% 2-Mercaptoethanol, 60 mM Tris-HCl pH 6.8, 2% SDS, 10 % Glycerol, 0.001 % bromophenol blue) at 95°C / 5 min. Samples were separated in the cold on 4-20 % SDS-PAGE and blotted for 1 h to nitrocellulose (pore size of 0.45 um), using: wet transfer in the cold. Blot was blocked with 5 % milk for: 2h/RT with agitation. Blot was incubated in the primary antibody at a dilution of 1: 1000 ON/4C with agitation in TBS-T. The antibody solution was decanted and the blot was rinsed briefly twice, then washed once for 15 min and 3 times for 5 min in TBS-T at RT with agitation. Blot was incubated in matching secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated) diluted to 1: 10 000 in 5% milk in TBS-T for 1h/RT with agitation. The blot was washed as above and developed with a following chemiluminescent detection reagent. Exposure time was 30 sec.
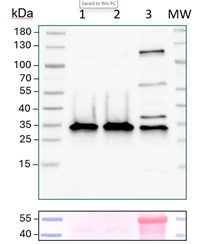
Antibody has been tested on the following mutants: Δ At3g08560 and Δ At4g11150.Triple mutant is lethal. Therefore vacuolar fraction has beenused for confirmation of reactivity of this antibody.
*The difference in loading amounts between the Cell Lysate (CL) and the vacuole samples is necessary because the protein concentration in the isolated vacuoles is very low. To ensure a detectable V-ATPase signal in the cellular fraction, I used 15 ug/well of CL protein. However, to obtain 15 ug of vacuolar protein I would need a large amount of sample, and purifying vacuoles is a long and complex procedure, so these samples are super valuable and limited.
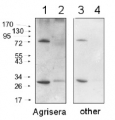
Courtesy of Dr. Lucia Borniego, Department of Biology, Indiana University Bloomington, USA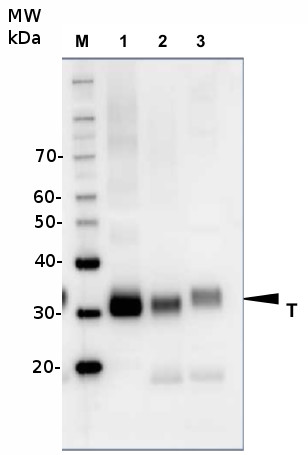 10 µg of total protein from samples such as Arabidopsis thaliana leaf (1) , Hordeum vulgare leaf (2), Zea mays leaf (3) were extracted with Protein Extraction Buffer PEB (AS08 300). Samples were diluted with 1X sample buffer (NuPAGE LDS sample buffer (Invitrogen) supplemented with 50 mM DTT and heat at 70°C for 5 min and keept on ice before loading. Protein samples were separated on 4-12% Bolt Plus gels, LDS-PAGE and blotted for 70 minutes to PVDF using tank transfer. Blots were blocked immediately following transfer in 2% blocking reagent or 5% non-fat milk dissolved in 20 mM Tris, 137 mM sodium chloride pH 7.6 with 0.1% (v/v) Tween-20 (TBS-T) for 1h at room temperature with agitation. Blots were incubated in the primary antibody at a dilution of 1: 5 000 (in blocking reagent) for 1h at room temperature with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, and then washed 1x15 min and 3x5 min with TBS-T at room temperature with agitation. Blots were incubated in secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated, recommended secondary antibody AS09 602, Agrisera) diluted to 1:25 000 in blocking reagent for 1h at room temperature with agitation. The blots were washed as above. The blot was developed for 5 min with chemiluminescent detection reagent, according the manufacturers instructions. Images of the blots were obtained using a CCD imager (VersaDoc MP 4000) and Quantity One software (Bio-Rad). Exposure time was 30 seconds.
10 µg of total protein from samples such as Arabidopsis thaliana leaf (1) , Hordeum vulgare leaf (2), Zea mays leaf (3) were extracted with Protein Extraction Buffer PEB (AS08 300). Samples were diluted with 1X sample buffer (NuPAGE LDS sample buffer (Invitrogen) supplemented with 50 mM DTT and heat at 70°C for 5 min and keept on ice before loading. Protein samples were separated on 4-12% Bolt Plus gels, LDS-PAGE and blotted for 70 minutes to PVDF using tank transfer. Blots were blocked immediately following transfer in 2% blocking reagent or 5% non-fat milk dissolved in 20 mM Tris, 137 mM sodium chloride pH 7.6 with 0.1% (v/v) Tween-20 (TBS-T) for 1h at room temperature with agitation. Blots were incubated in the primary antibody at a dilution of 1: 5 000 (in blocking reagent) for 1h at room temperature with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, and then washed 1x15 min and 3x5 min with TBS-T at room temperature with agitation. Blots were incubated in secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated, recommended secondary antibody AS09 602, Agrisera) diluted to 1:25 000 in blocking reagent for 1h at room temperature with agitation. The blots were washed as above. The blot was developed for 5 min with chemiluminescent detection reagent, according the manufacturers instructions. Images of the blots were obtained using a CCD imager (VersaDoc MP 4000) and Quantity One software (Bio-Rad). Exposure time was 30 seconds.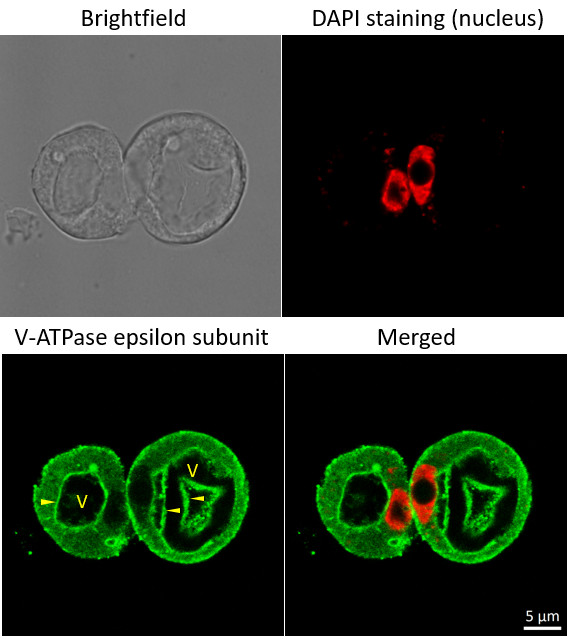
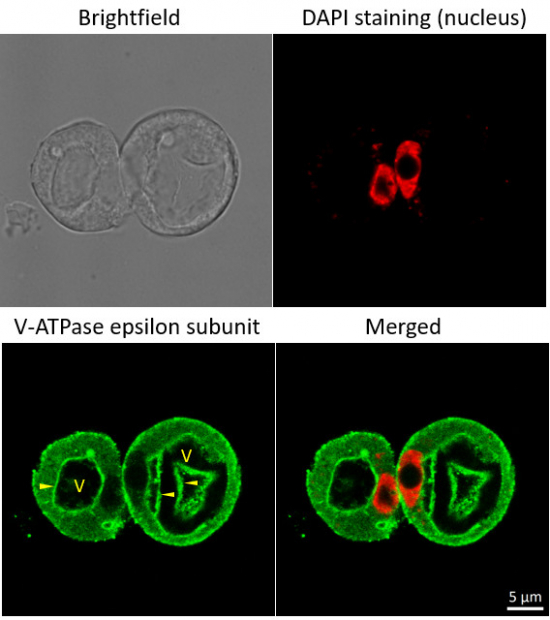
Immunofluorescent localization of V-ATPase epsilon subunit of tonoplast H+ATPase in suspension culture of Oryza sativa ssp. japonica cv. 'Unggi 9', using goat anti-V-ATPase, epsilon subunit of tonoplast antibodies (AS07 213) and donkey anti-rabbit IgG, DyLight® 488 conjugated (AS10 1165, Agrisera). Vacuolar membrane, tonoplast, is highlighted by yellow arrowheads. DAPI staining of nuclei is pseudocolored red.
Method
Material: Suspension cultures of Oryza sativa ssp. japonica cv. 'Unggi 9
Fixation: Packed cell volume to fixer ratio: 250 µl : 5ml
Fixer composition and buffer: 4% (w/v) paraformaldehyde (freshly prepared as 8% stock and 0.2 µm filtered) 0.01% (v/v) Triton-X100 in Phosphate Buffered Saline (PBS), pH 7.4 (2x stock, 0.2 µm filtered)Container and method: in 6 cm Petri dish, gentle shaking at room temperature (RT) Duration: 40 min
Hydrophilization: No
Cell wall digestion: Yes Packed cell volume to enzyme ratio: 100ul : 2ml Enzyme composition: 1% (A) 1.2% (R) Cellulase (chromatically purified, powder, Worthington) 1% (A) 1.2% (R) Pectinase (protease free, liquid, Sigma) Buffer: 0.5% (w/v) MES buffer, pH 5.6 Container and method: in 2 ml microfuge tube by rolling at room temperature (RT)
Duration: 60 min
Membrane permeabilization: Triton-X100 (0.35%) 7 mins/RT
Antigen retrieval: No
Blocking buffer: Fish gelatin (5% v/v) Washing buffer: PBS
Primary antibody dilution and incubation time: 1:300, ON/4°C
Secondary antibody: donkey anti-rabbit IgG, DyLight® 488 conjugated (AS10 1165, Agrisera), 1:600, 1h/RT
Co-staining of the nucleus (DAPI):
Cell wall and nucleus staining: 100 ng/ml DAPI
Courtesy of Dr. Ferhan Ayaydin, Hungarian Centre of Excellence for Molecular Medicine (HCEMM), Szeged, Hungary.Application examples: 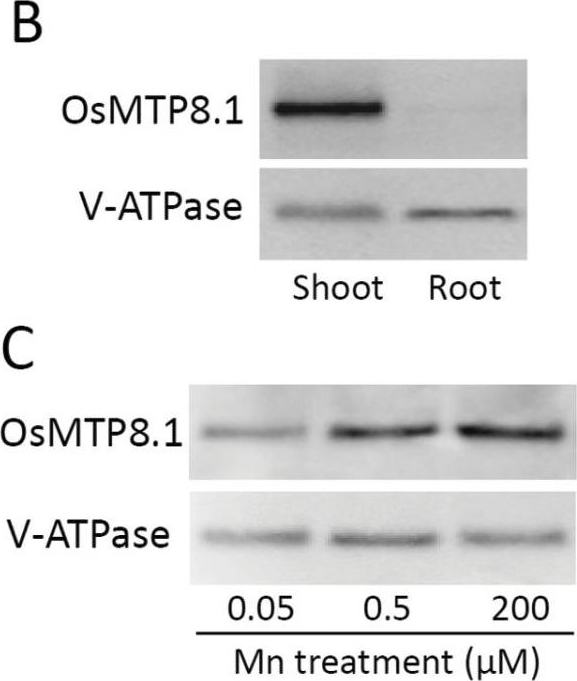
Reactant: Oryza sativa (Asian rice)
Application: Western Blotting
Pudmed ID: 23963678
Journal: J Exp Bot
Figure Number: 3B,C
Published Date: 2013-11-01
First Author: Chen, Z., Fujii, Y., et al.
Impact Factor: 6.088
Open PublicationExpression pattern of OsMTP8.1. (A) Quantitative real-time RT–PCR analysis of OsMTP8.1 expression in rice shoots and roots grown in different Mn concentrations. Plants were hydroponically grown for 12 d and then for 6 d in a solution containing varying concentrations of Mn (0.05, 0.5, and 200 µM). Histone H3 was used as an internal control. Expression relative to the shoots in the presence of 0.5 µM Mn is shown. Data represent the mean ±SD (n=3). Different letters indicate a significant difference at P < 0.05 using Tukey’s test. (B) Western blot analysis of OsMTP8.1 in shoots and roots. The tonoplast marker protein V-ATPase was detected using a specific antibody. (C) Western blot analysis of OsMTP8.1 in shoots grown in the presence of varying concentrations of Mn (0.05, 0.5, and 200 µM). (D, E) Immunostaining of the leaf blades of the OsMTP8.1 promoter–GFP transgenic line (D) and wild-type rice (E). Scale bars=50 µm.
Reactant: Oryza sativa (Asian rice)
Application: Western Blotting
Pudmed ID: 23963678
Journal: J Exp Bot
Figure Number: 4C,D
Published Date: 2013-11-01
First Author: Chen, Z., Fujii, Y., et al.
Impact Factor: 6.088
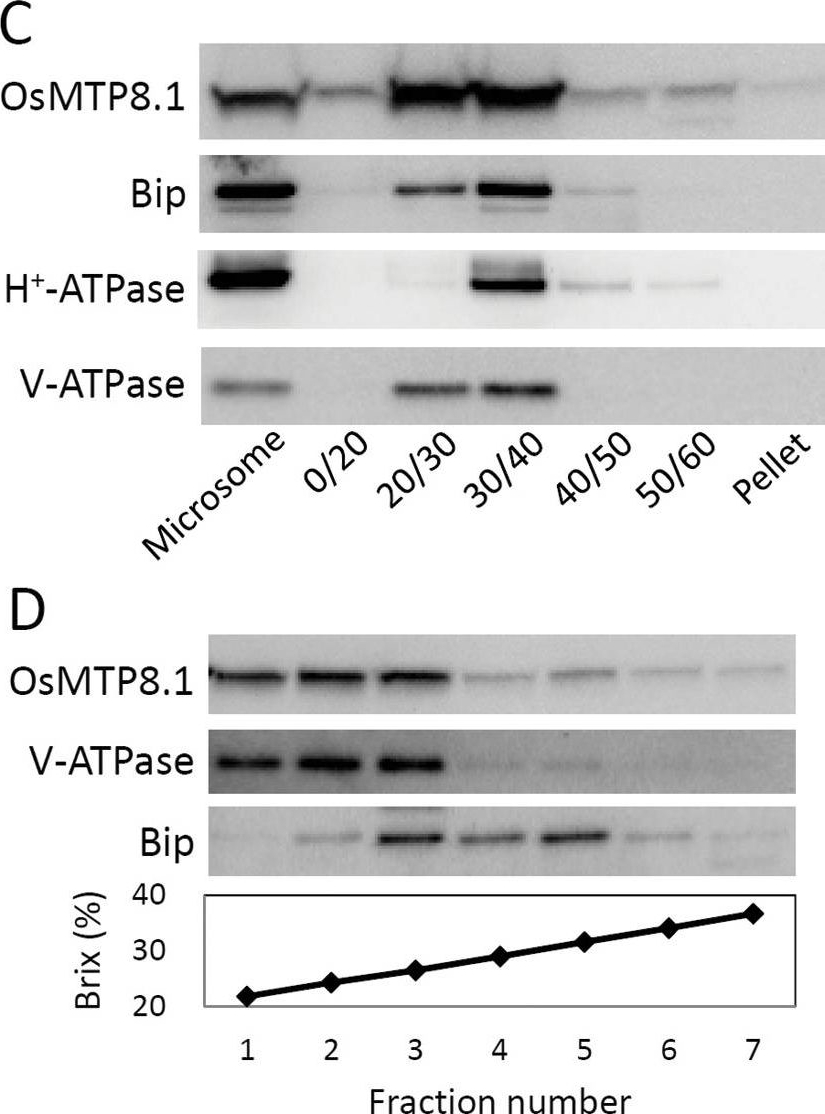
Open PublicationSubcellular localization of OsMTP8.1. (A, B) Localization of GFP (A) and the OsMTP8.1:GFP fusion protein (B) transiently expressed in onion epidermal cells. A magnified view of the image surrounding the nucleus is shown in (B). Expression was monitored 12h after transformation. Scale bars=100 µm. (C) Western blot analysis of OsMTP8.1 in shoot membrane fractions prepared using a discontinuous sucrose density gradient. Antibodies to marker proteins of the ER (anti-Bip), plasma membrane (anti-H+-ATPase), and tonoplast (anti-V-ATPase) were used to probe the blots. (D) Western blot analysis of OsMTP8.1 in microsome fractions prepared in the presence of Mg2+. Brix=1g of sucrose in 100g of solution.
Reactant: Oryza sativa (Asian rice)
Application: Western Blotting
Pudmed ID: 29479780
Journal: Plant Biotechnol J
Figure Number: 1C
Published Date: 2018-10-01
First Author: Deng, F., Yamaji, N., et al.
Impact Factor: 8.66
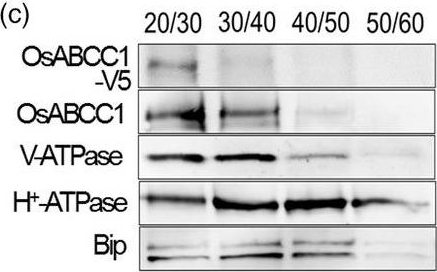
Open PublicationDevelopment of transgenic rice with reduced As accumulation in the grains using tissue?specific expression of OsABCC1, ??ECS and ScYCF1. (a) Analysis of the tissue?specific expression of the OsRCc3 promoter by immunostaining OsRCc3 promoter::GUS transgenic plants (RCc3p::GUS) using a GUS antibody. The internode section denoted by broken lines is magnified in the inset surrounded by yellow solid lines. Bar = 100 ?m. (b) Maps of vectors carrying RCc3p?A (RCc3 pro::OsABCC1?V5), RCc3p?AE (RCc3 pro::OsABCC1?V5, ZmUBI pro::??ECS) or RCc3p?AEY (RCc3 pro::OsABCC1?V5, ZmUBI pro::??ECS, RCc3 pro::ScYCF1). (c) Subcellular localization of OsABCC1?V5 in the roots of RCc3p?AEY plants using a sucrose density gradient analysis. Anti?V5, anti?OsABCC1, anti?V?ATPase (tonoplast marker), anti?H+?ATPase (plasma membrane marker) and Bip (ER marker) were used to assess protein abundance in the different compartments. The signal of OsABCC1?V5 corresponded to the signals of OsABCC1 and V?ATPase. (d) Arsenic accumulation in brown rice harvested from T3 transgenic plants transformed with RCc3p?A, RCc3p?AE, RCc3p?AEY, UBIp?A, UBIp?AE or UBIp?AEY. The As concentration was analysed in brown rice from plants grown in soil with a typical basal level of As. The values are means and standard errors (n = 5 plants). Different letters indicate significantly different means (Tukey's multiple comparison analysis, P ? 0.05).
- Additional Information
-
Additional information: Cellular [compartment marker] of tonoplast membrane.
This product can be sold containing ProClin if required.Additional information (application): V-ATPase is very sensitive for the redox of the SDS buffer. We recommend using at least 50-100 mM DTT freshly prepared before handling the sample.
Immunostaining protocol using V-ATPase antibodies can be found here.
- Background
-
Background: Plant vacuole V-ATPase is responsible for energization of transport of ions and metabolites, and acts as well 'house-keeping' and as a stress response enzyme. V-ATPase is a multi-subunit enzyme composed of a membrane sector and a cytosolic catalytic sector. It is related to the FoF1 ATP synthase.
Alternative protein names: Vacuolar proton pump subunit E, Protein EMBRYO DEFECTIVE 2448 - Product Citations
-
Selected references: Holzner et al. (2026). The chloroplast ionome shines light on the dynamics of organellar iron homeostasis. Plant Cell. 2026 Jan 29:koag017. doi: 10.1093/plcell/koag017.
Ramakrishna et al. (2025). Elemental cryo-imaging reveals SOS1-dependent vacuolar sodium accumulation. Nature. 2025 Jan;637(8048):1228-1233. doi: 10.1038/s41586-024-08403-y. Epub 2025 Jan 15.
Salazar et al. (2024). SOS1 tonoplast neo-localization and the RGG protein SALTY are important in the extreme salinity tolerance of Salicornia bigelovii. Nat Commun. 2024 May 20;15(1):4279.doi: 10.1038/s41467-024-48595-5.
Collins et al. (2020). EPSIN1 Modulates the Plasma Membrane Abundance of FLAGELLIN SENSING2 for Effective Immune Responses . Plant Physiol. 2020 Feb 24. pii: pp.01172.2019. doi: 10.1104/pp.19.01172
Skalicky et al. (2021) Auxin Metabolite Profiling in Isolated and Intact Plant Nuclei. Int J Mol Sci. 2021 Nov 16;22(22):12369. doi: 10.3390/ijms222212369. PMID: 34830250; PMCID: PMC8620152.
Roustan et al. (2020). Protein sorting into protein bodies during barley endosperm development is putatively regulated by cytoskeleton members, MVBs and the HvSNF7s. Sci Rep. 2020 Feb 5;10(1):1864. doi: 10.1038/s41598-020-58740-x.
Prinsi et al. (2020). Root Proteomic Analysis of Two Grapevine Rootstock Genotypes Showing Different Susceptibility to Salt Stress. Int J Mol Sci. 2020 Feb 6;21(3). pii: E1076. doi: 10.3390/ijms21031076.
Kuang et al. (2019). Quantitative Proteome Analysis Reveals Changes in the Protein Landscape During Grape Berry Development With a Focus on Vacuolar Transport Proteins. Front Plant Sci. 2019 May 15;10:641. doi: 10.3389/fpls.2019.00641. eCollection 2019.
Pertl-Obermeyer et al. (2018). Dissecting the subcellular membrane proteome reveals enrichment of H+ (co-)transporters and vesicle trafficking proteins in acidic zones of Chara internodal cells. PLoS One. 2018 Aug 29;13(8):e0201480. doi: 10.1371/journal.pone.0201480.
Migocka et al. (2018). Cucumber metal tolerance protein 7 (CsMTP7) is involved in the accumulation of Fe in mitochondria under Fe excess. Plant J. 2018 Jun 22. doi: 10.1111/tpj.14006.
Zhu et al. (2018). A comprehensive proteomic analysis of elaioplasts from citrus fruits reveals insights into elaioplast biogenesis and function. Hortic Res. 2018 Feb 7;5:6. doi: 10.1038/s41438-017-0014-x.
Lynch et al. (2017). Multifaceted plant responses to circumvent Phe hyperaccumulation by downregulation of flux through the shikimate pathway and by vacuolar Phe sequestration. Plant J. 2017 Dec;92(5):939-950. doi: 10.1111/tpj.13730.
Nagel et al. (2017). Arabidopsis SH3P2 is an ubiquitin-binding protein that functions together with ESCRT-I and the deubiquitylating enzyme AMSH3. Proc Natl Acad Sci U S A. 2017 Aug 7. pii: 201710866. doi: 10.1073/pnas.1710866114.
Vera-Estrella et al. (2017). Cadmium and zinc activate adaptive mechanisms in Nicotiana tabacum similar to those observed in metal tolerant plants. Planta. 2017 Apr 28. doi: 10.1007/s00425-017-2700-1.
Xing et al. (2016). Proteome Profile of Starch Granules Purified from Rice (Oryza sativa) Endosperm. PLoS One. 2016 Dec 19;11(12):e0168467. doi: 10.1371/journal.pone.0168467.
LaMontagne et al. (2016). Isolation of Microsomal Membrane Proteins from Arabidopsis thaliana. Curr. Protoc. Plant Biol. 1:217-234. doi: 10.1002/cppb.20020.
Barkla et al. (2016). Single-cell-type quantitative proteomic and ionomic analysis of epidermal bladder cells from the halophyte model plant Mesembryanthemum crystallinum to identify salt-responsive proteins. BMC Plant Biol. 2016 May 10;16(1):110. doi: 10.1186/s12870-016-0797-1.
Liu et al. (2016). iTRAQ-based quantitative proteomic analysis reveals the role of the tonoplast in fruit senescence. J Proteomics. 2016 Sep 2;146:80-9. doi: 10.1016/j.jprot.2016.06.031.
Wattelet-Boyer et al. (2016). Enrichment of hydroxylated C24- and C26-acyl- chain sphingolipids mediates PIN2 apical sorting at trans-Golgi network subdomains. Nat Commun. 2016 Sep 29;7:12788. doi: 10.1038/ncomms12788.
Jiskrova et al. (2016). Extra- and intracellular distribution of cytokinins in the leaves of monocots and dicots. N Biotechnol. 2016 Jan 8. pii: S1871-6784(16)00002-9. doi: 10.1016/j.nbt.2015.12.010
Lang et al. (2011).Simultaneous isolation of pure and intact chloroplasts and mitochondria from moss as the basis for sub-cellular proteomics. Plant Cell Rep. 2011 Feb;30(2):205-15.doi: 10.1007/s00299-010-0935-4. - Protocols
-
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extractsImmunostaining of cytoskeletal structures in guard cells of Commelina communis
Fixation:
Peel the epidermis and immediately transfer to the fixative:
1.PME buffer (50mM PIPES PH 6.9, 5mM EGTA, 2mM MgSO4) with 0.05% Triton X-100, 0.25% DMSO, 1mM PMSF, 200-500 µM MBS. (MBS is not stable in solution, it is thus important to prepare it just before use, add the MBS drop by drop to the solution while stirring, because it may precipitate, MBS is needed only to stain for actin filaments).
2.Incubate 5 minutes.
3.Transfer the peels to 3% PFA (paraformaldehyde) in PME buffer with 0.05% Triton X-100, 0.25% DMSO, 50 µM PMSF.
4.Incubate 1hour.
5.Wash twice for 5 minutes with PBS.Permeabilization:
1.Put the epidermal peels between two microscope glass and freeze in liquid nitrogen for 1-2 minutes.
2.Place the glass between two aluminum blocks cooled to –80oC and punch from above gently.
3.Open the two glass while frozen, and slide the epidermal peels into a solution of PBS + 1% Triton X-100, incubate 1.5 hours.
4.Wash with PBS.Staining:
1.Block over night in PBS with 0.05% Triton X-100 and 1% BSA at 4oC.
2.Incubate 1-1.5 hours in the presence of the first antibody diluted in PBS + 0.05% Triton-X100 and 1% BSA.
3.Wash 3X30 minutes in PBS.
4.Incubate in the presence of the secondary antibody as above.
5.Wash as above.
6.Mount in Elvanol.Courtesy Dr Einat Sadot , Department of ornamental Horticulture, ARO Volcani center, Israel
- Reviews:
-
Praveen Kumar | 2014-05-30I used it detect the microsome fraction from the ARabidopsis Col-0 background. It worked great at 1:20000 dilution.Gerhard Obermeyer | 2009-11-18An excellent antibody against subunit E of the V-ATPase. The antibody recognised only one protein band in Western blots at ca. 30 kDa in a microsomal fraction prepared from Lilium pollen grains. Anti-VHA-E was diluted 1:2,500 and an anti-rabbit IgG AP-conjugated (1:5,000) was used for detection.| 2009-07-21This antibody worked very well in the fern Pteris vittata. I used the recommended dilution of 1:1000 and had no problems.Bronwyn Barkla | 2008-11-17Excellent antibody for tonoplast. Recognizes VHA-E subunit from many different species.Highly recommended.