1
Anti-AOX1/2 | Plant alternative oxidase 1 and 2
AS04 054 | Clonality: Polyclonal | Host: Rabbit | Reactivity: A. thaliana, B. nana, B. napus, B. oleracea, B. vulgaris, E. vaginatum, H. vulgare, L. luteus, N.tabacum, O. sativa, P.abies, P. sativum, S.tuberosum and T. aestivum, conifers and P. patens cellular[compartment marker] of mitochondrial inner membrane
Benefits of using this antibody
- Product Info
-
Immunogen: KLH-conjugated synthetic peptide derived from fully conserved C-terminal consensus motif from plant AOX isoforms including Arabidopsis thaliana AOX1A. UniProt: Q39219, TAIR: At3g22370, AOX1B UniProt: O23913, TAIR:AT3G22360, AOX1C UniProt: O22048, TAIR: AT3G27620, and AOX2, UniProt: O22049, TAIR: AT5G64210, Solanum lycopersicum UniProt: Q7XBG9, Oryza sativa UniProt: Q7XT33, AOX1D, TAIR: AT1G32350
Host: Rabbit Clonality: Polyclonal Purity: Serum Format: Lyophilized Quantity: 50 µl Reconstitution: For reconstitution, add 50 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Immunolocalization (IL), Western blot (WB) Recommended dilution: 1 : 750 (IL), 1 : 1000 for 10-20 µg of mitochondrial protein/lane detection (WB) Expected | apparent MW: 36-40 | 36-40 for Arabidopsis thaliana - Reactivity
-
Confirmed reactivity: Arabidopsis thaliana, Betula nana, Beta vulgaris, Brassica napus, Brassica oleracea, Kandelia candel, Eriphorum vaginatum, Glycine max, Hordeum vulgare, Lupinus luteus, Nicotiana tabacum, Oryza sativa, Picea abies, Pisum sativum, Poa annua, Robinia pseudoacacia, Rosa hybrida, Solanum lycopersicum, Solanum tuberosum, Symplocarpus renifolius, Physcomitrium patens, Tigriopus californicus, Triticum aestivum, Zea mays
Predicted reactivity: Aegilops tauschii, Brachypodium distachyon, Capsella rubella, Citrus sinensis, Citrus clementina, Corylus heterophylla, Crocus sativus, Cucumis sativus, Daucus carota, Hypericum perforatum, Lotus japonicus, Malus x domestica, Medicago truncatula, Medicago sativa, Mangifera indica, Naegleria gruberi (amoeba), Nelumbo nucifera, Nicotiana benthamiana, Oryza brachyantha, Populus tremula, Picea sitchensis, Pinus pinaster, Pyrus communis, Saccharum officinarum, Sauromatum venosum, Selaginella moellendorffii, Sorghum bicolor, Spinacia oleracea, Tetrahymena thermophila, Vigna radiata, Vigna unguiculata, Vitis vinifera
Species of your interest not listed? Contact usNot reactive in: Candidia albicans, Chlamydomonas reinhardtii (use an antibody to algal AOX1, AS06 152), Stomolophus sp2
- Application Examples
-
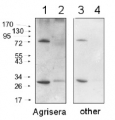
25 μg of Arabidopsis thaliana mitochondrial wild type fraction (1) mitochondrial fraction from a mutant with increased AOX level (2), total wild type leaf extract (3), total leaf extract from AOX overproducing mutant (4) were separated on 10% gel and blotted on nitrocellulose membrane using wet transfer (0.22% CAPS, pH 11). Filters where blocked (1.5h) in 5% milk in TBST (1X TBS, 0,1% Tween 20), incubated with 1: 1000 anti-AOX polyclonal antibodies (2h in TBST) followed by 1 h incubation with 1: 50 000 Agrisera secondary anti-rabbit HRP-coupled antibodies (AS09 602) and visualized with chemiluminescent detection reagent, on Kodak autoradiography film for 15-60 s. Mitochondria were isolated as described by Urantowka et al. (Plant Mol Biol, 2005, 59:239-52). Mitochondrial pellets were suspended in 1X Laemmli buffer (5% beta-mercaptoetanol, 3.7% glycerol, 1.1% SDS, 23 mM Tris- HCl pH 6.8, 0.01% bromophenol blue), heated (95°C, 5 min.) and centrifuged (13 000rpm, 1 min.). Leaf extracts were prepared as described by Martinez-Garcia et al. (Plant J., 1999, 20:251-7).
Courtesy Dr. Janusz Piechota, Wrocław University, Poland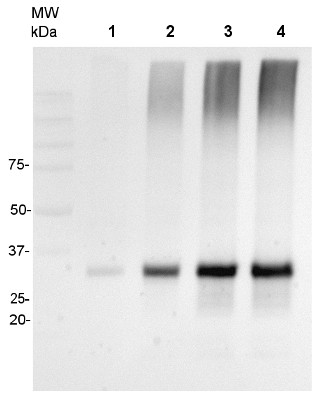
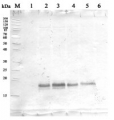
20 μg of mitochondrial protein isolated from 2-week-old Arabidopsis thaliana seedlings (Smakowska et al., 2016) extracted with a buffer containing urea, thiourea, CHAPS and Triton X-100 (Heidorn-Czarna et al., 2018) were denaturated with Laemmli buffer at 95°C for 5 min and separated on 12% SDS-PAGE. Wild-type grown at 22°C (1), mutant grown at 22°C (2), wild-type grown at 30°C (3), mutant grown at 30°C.
Afterwards the gel was blotted for 1.5h to nitrocellulose membrane using wet-transfer. Blot was blocked with 5% milk in TBS-T at 4°C/ON with agitation. Blot was incubated in the primary antibody (anti-AOX1/2, AS04 054) at a dilution 1:1000 in 5% milk in TBS-T for 1.5h /RT with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, then washed once for 15 min and 2 times for min in TBS-T at RT with agitation.Blot was incubated in Agrisera matching secondary antibody (goat anti-rabbit IgG, HRP-conjugated, AS09 602) diluted to 1:20 000 in 5% milk in TBS-T for 1h/RT with agitation. The blot was washed as above and developed with chemiluminescence using GBox imager (Syngene).
Courtesy Dr. Małgorzata Heidorn-Czarna, University of Wrocław, Poland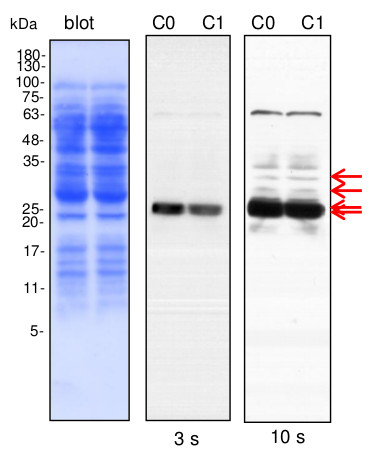
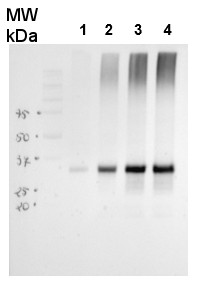
Lines C0, C1- 10 µg of cauliflower mitochondrial proteins (C0- controls; C1- plants grown in mild drought conditions) isolated as described by Rurek et al., 2015 (doi:10.1016/j.bbabio.2015.01.005) were separated by 12% SDS- PAGE and electroblotted in semi-dry conditions (Towbin buffer) to Immobilon-P membrane (Millipore). Blots were CBB R 250 briefly stained, destained, wet-scanned and after completed destaining, they were blocked in 5% skimmed milk (dissolved in PBS-T containing 0.1% Tween 20) in 1h, RT. Primary antisera (at 1: 1000, diluted in 2% skimmed milk in PBS-T) were bound by overnight incubation of blots at +4 O C. After blot washing (2 times quick, 2 times of 5 min, and 10 min at the end), secondary goat anti-rabbit IgGs, HRP- conjugated (Agrisera, AS09 602; at 1: 50 000, diluted in 2% milk/ PBS-T) were bound in 1 h, RT. Blots were washed (as above) with copious amounts of PBS-T and chemiluminescence signals acquired by using chemiluminescent detection reagents on RTG film between 3 s and 2 min (periods of the given image acquisition were indicated).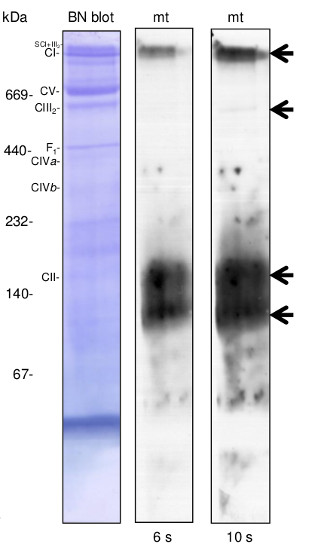
100 µg of cauliflower mitochondria were pelleted and proteins were digitonin solubilised (30 min at 4°C) at the detergent: protein ratio 4:1 (g:g) using ACA 750 buffer. Unsolubilised material was further pelleted and supernatant after complementation with Serva Blue was loaded onto 4.5-16% gradient BN gel. After separation, protein complexes in the gel were denatured and reduced (in the presence of SDS and 2-mercaptoethanol) and then they were electroblotted and immunodetected essentially in the same manner as it was indicated for SDS-PAGE blots. Four complexes containing alternative oxidase were detected (the most abundant ca.150 and 120 kDa). This data is very similar to the one obtained for green tissue mitochondria of Arabidopsis and Medicago (see Gelmap project; https://gelmap.de/). Mobility of known OHPHOS complexes (complex I, II, III, IV and ATP synthase= complex V) was additionally indicated.
Courtesy Dr. Michał Rurek, Department of Molecular and Cellular Biology, Institute of Molecular Biology and Biotechnology, Faculty of Biology, Adam Mickiewicz University in Poznań, PolandApplication examples: 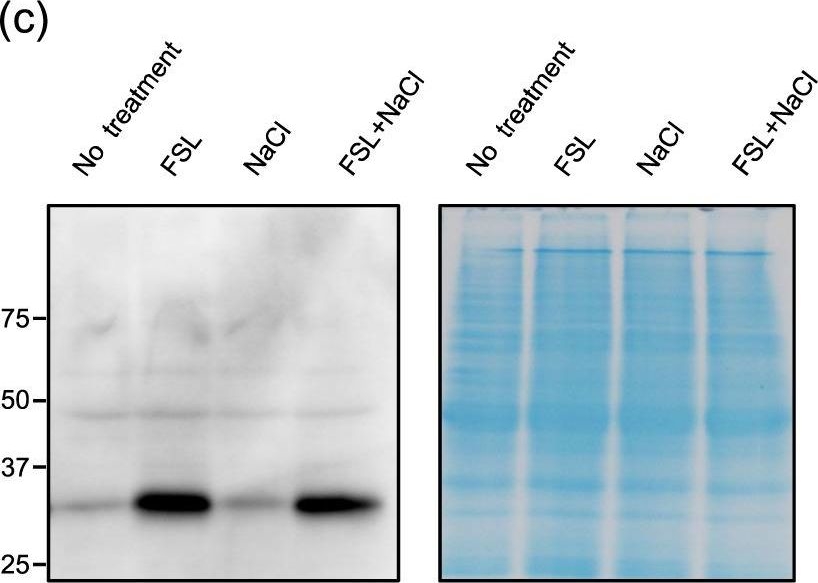
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 32457324
Journal: Sci Rep
Figure Number: 2C
Published Date: 2020-05-26
First Author: Sako, K., Futamura, Y., et al.
Impact Factor: 4.13
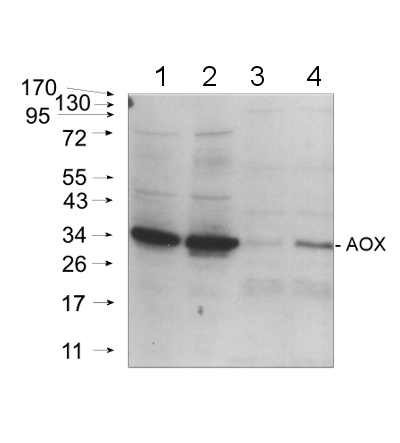
Open PublicationExpression profile of genes up-regulated by both FSL0260 treatment and salinity stress. (a) Cellular component gene ontology of up-regulated genes by FSL0260 treatment. (b) Relative expression levels of AtNDB4 and AtAOX1a genes during salinity-stress treatment for 0 and 2?h with or without 20?µM FSL0260. Expression level of plants treated with DMSO was set as 1. 18S rRNA was used as an internal standard. Error bars represent the mean ± SE (n?=?3). Statistical significance was determined by ANOVA, followed by post-hoc Tukey’s tests. Means that differed significantly (P?<?0.05) are indicated by different letters. (c) Immunoblot of the AOX (35?kDa) proteins (left). Coomassie blue-stained gel showing control loading (right). Total proteins were extracted from seedlings treated with 0 or 20?µM FSL0260 for 24?h and with or without subsequent treatment of 100?mM NaCl for 6?h. DMSO was used as a negative control. Immunoblot analysis was performed using an anti-AOX1/2 antibody. (d) The signal intensity of AOX1/2. DMSO treatment was taken as 1. Error bars represent the mean ± SE (n?=?3). Statistical significance was determined by ANOVA, followed by post-hoc Tukey’s tests. Means that differed significantly (P?<?0.05) are indicated by different letters.
- Additional Information
-
Additional information: Mitochondrion inner membrane marker. Possibly in the inner surface of the inner mitochondrial membrane.
Protocol for a plant mitochondria preparation can be found here.
In protein samples which are older than few months AOX enzyme can undergo intensive dimerization. Such preparations should not be used to work with this antibody.Additional information (application): According to Konert et al. (2015) AOX antibody is recognizing AOX1A and AOX1D.
This product can be sold containing ProClin if requested. - Background
-
Background: Alternative oxidases (AOX) are quinol oxidases located in the inner mitochondrial membrane of plants. They function as terminal oxidases in the alternate electron transport pathway, oxidizing ubiquinone to reduce oxygen to water.
- Product Citations
-
Selected references: Punkkinen et al. (2025). Mitochondria affect photosynthesis through altered tissue levels of O₂.Plant Physiol. 2025 Dec 12:kiaf648. doi: 10.1093/plphys/kiaf648.
Keresztes et al. (2025). Alternative oxidase activity and vacuolar intrusion of mitochondria represent a delayed mitophagy associated with the chilling stress in Haberlea rhodopensis. Plant Stress, October 2025.
Iglesias-Sanchez et al. (2025).Activation of Alternative Oxidase Ensures Carbon Supply for Ethylene and Carotenoid Biosynthesis During Tomato Fruit Ripening. Plant Physiol. 2025 Oct 31;199(3):kiaf516. doi: 10.1093/plphys/kiaf516.
Shi et al. (2025). Quantitative Analyses of Programmed Cell Death (PCD) Relevant Indicators in Postharvest Vegetable. Future Postharvest and Food. doi.org/10.1002/fpf2.70027.
Dong et al. (2025). SGAM1 orchestrates salt tolerance by balancing mitochondrial translation and ROS homeostasis in Arabidopsis. Plant J. 2025 Jul;123(1):e70322.doi: 10.1111/tpj.70322.
Hoa et al. (2024). Proteomic analysis on symbiotic differentiation of mitochondria in soybean nodules. Comparative Study Plant Cell Physiol. 2004 Mar;45(3):300-8. doi: 10.1093/pcp/pch035.
Soria et al. (2024).Functional resilience: An active oxidative phosphorylation system prevails amid foreign proteins in holoparasitic plants. Current Plant Biology Volume 37, March 2024, 100322.
Chen et al. (2023) The pentatricopeptide repeat protein EMP601 functions in maize seed development by affecting RNA editing of mitochondrial transcript ccmC, https://doi.org/10.1016/j.cj.2023.03.004.
Rodrigues et al. (2023). Germination of Pisum sativum L. Seeds Is Associated with the Alternative Respiratory Pathway. Biology (Basel). 2023 Oct 9;12(10):1318.doi: 10.3390/biology12101318.
Wang, Y., Ye, H., Gao, K. et al. The opening of mitochondrial permeability transition pore (mPTP) and the inhibition of electron transfer chain (ETC) induce mitophagy in wheat roots under waterlogging stress. Protoplasma 260, 1179–1191 (2023).
Vainonen et al. (2023) Poly(ADP-ribose)-binding protein RCD1 is a plant PARylation reader regulated by Photoregulatory Protein Kinases. Commun Biol. 2023 Apr 19;6(1):429. doi: 10.1038/s42003-023-04794-2
Nguyen et al. (2022). MISF2 Encodes an Essential Mitochondrial Splicing Cofactor Required for nad2 mRNA Processing and Embryo Development in Arabidopsis thaliana. Int J Mol Sci. 2022 Feb 28;23(5):2670. doi: 10.3390/ijms23052670. PMID: 35269810; PMCID: PMC8910670.
Brito et al. (2022) The role of the electron-transfer flavoprotein: ubiquinone oxidoreductase following carbohydrate starvation in Arabidopsis cell cultures. Plant Cell Rep. 2022 Jan 15. doi: 10.1007/s00299-021-02822-1. Epub ahead of print. PMID: 35031834.
Pascual et al (2021). ACONITASE 3 is part of the ANAC017 transcription factor-dependent mitochondrial dysfunction response, Plant Physiology, 2021;, kiab225, https://doi.org/10.1093/plphys/kiab225
Challabathula et al. (2021) Differential modulation of photosynthesis, ROS and antioxidant enzyme activities in stress-sensitive and -tolerant rice cultivars during salinity and drought upon restriction of COX and AOX pathways of mitochondrial oxidative electron transport, Journal of Plant Physiology,Volume 268,2022,153583, ISSN 0176-1617,https://doi.org/10.1016/j.jplph.2021.153583.
Oh et al. (2021) Alternative oxidase (AOX) 1a and 1d limit proline-induced oxidative stress and aid salinity recovery in Arabidopsis. Plant Physiol. 2021 Dec 17:kiab578. doi: 10.1093/plphys/kiab578. Epub ahead of print. PMID: 34919733.
Pavlovic & Kocab. (2021) Alternative oxidase (AOX) in the carnivorous pitcher plants of the genus Nepenthes: what is it good for? Ann Bot. 2021 Dec 18:mcab151. doi: 10.1093/aob/mcab151. Epub ahead of print. PMID: 34922341.
Makino et al. (2020). Induction of Terminal Oxidases of Electron Transport Chain in Broccoli Heads Under Controlled Atmosphere Storage. Foods, 9 (4)
Marchetti et al. (2020). Mitochondrial Pentatricopeptide Repeat Protein, EMB2794, Plays a Pivotal Role in NADH Dehydrogenase Subunit nad2 mRNA Maturation in Arabidopsis Thaliana. Plant Cell Physiol DOI: 10.1093/pcp/pcaa028
Garmash et al. (2020). Altered levels of AOX1a expression result in changes in metabolic pathways in Arabidopsis thaliana plants acclimated to low dose rates of ultraviolet B radiation. Plant Sci. 2020 Feb;291:110332. doi: 10.1016/j.plantsci.2019.110332.
Kuang et al. (2019). Quantitative Proteome Analysis Reveals Changes in the Protein Landscape During Grape Berry Development With a Focus on Vacuolar Transport Proteins. Front Plant Sci. 2019 May 15;10:641. doi: 10.3389/fpls.2019.00641. eCollection 2019.
Tward et al. (2019). Identification of the alternative oxidase gene and its expression in the copepod Tigriopus californicus. Comp Biochem Physiol B Biochem Mol Biol. 2019 Feb;228:41-50. doi: 10.1016/j.cbpb.2018.11.003.
Rethore et al. (2019). Arabidopsis seedlings display a remarkable resilience under severe mineral starvation using their metabolic plasticity to remain self-sufficient for weeks. Plant J. 2019 Mar 22. doi: 10.1111/tpj.14325.
Luevano-Martínez et al. (2019). Mitochondrial alternative oxidase is determinant for growth and sporulation in the early diverging fungus Blastocladiella emersonii. Fungal Biology, Vol 123, Issue 1, 59-65.
Cordoba et al. (2019). Different Types of CA Domains Are Present in Complex I from Immature Seeds and Adult Plants in Arabidopsis thaliana. Plant Cell Physiol. 2019 Jan 22. doi: 10.1093/pcp/pcz011.
Czobor et al. (2019). Comparison of the response of alternative oxidase and uncoupling proteins to bacterial elicitor induced oxidative burst. PLoS One. 2019 Jan 10;14(1):e0210592. doi: 10.1371/journal.pone.0210592.
Hu et al. (2018). OsNDUFA9 encoding a mitochondrial complex I subunit is essential for embryo development and starch synthesis in rice. Plant Cell Rep. 2018 Dec;37(12):1667-1679. doi: 10.1007/s00299-018-2338-x.
Borovik and Grabelnych (2018). Mitochondrial alternative cyanide-resistant oxidase is involved in an increase of heat stress tolerance in spring wheat. J Plant Physiol. 2018 Dec;231:310-317. doi: 10.1016/j.jplph.2018.10.007.
Umekawa and Ito (2018). Thioredoxin o-mediated reduction of mitochondrial alternative oxidase in the thermogenic skunk cabbage Symplocarpus renifolius. J Biochem. 2018 Oct 5. doi: 10.1093/jb/mvy082.
Hu et al. (2018). OsNDUFA9 encoding a mitochondrial complex I subunit is essential for embryo development and starch synthesis in rice.Plant Cell Rep. 2018 Aug 27. doi: 10.1007/s00299-018-2338-x.
Zhu et al. (2018). Mitochondrial alternative oxidase-dependent autophagy involved in ethylene-mediated drought tolerance in Solanum lycopersicum. Plant Biotechnol J. 2018 May 4. doi: 10.1111/pbi.12939.
Garmash et al. (2017). Expression profiles of genes for mitochondrial respiratory energy-dissipating systems and antioxidant enzymes in wheat leaves during de-etiolation. J Plant Physiol. 2017 Aug;215:110-121. doi: 10.1016/j.jplph.2017.05.023.
Vishwakarma et al. (2016). A discrete role for alternative oxidase under hypoxia to increase nitric oxide and drive energy production. Free Radic Biol Med. 2018 Mar 28. pii: S0891-5849(18)30148-5. doi: 10.1016/j.freeradbiomed.2018.03.045.
Zhao et al. (2016). Nitrogen deprivation induces cross-tolerance of Poa annua callus to salt stress. Biologia Plantarum 60 (3): 543–554.
Solti et al. (2016). Does a voltage-sensitive outer envelope transport mechanism contributes to the chloroplast iron uptake? Planta. 2016 Dec;244(6):1303-1313. Epub 2016 Aug 19.
Zhang et al. (2016). A High Temperature-Dependent Mitochondrial Lipase EXTRA GLUME1 Promotes Floral Phenotypic Robustness against Temperature Fluctuation in Rice (Oryza sativa L.). PLoS Genet. 2016 Jul 1;12(7):e1006152. doi: 10.1371/journal.pgen.1006152. eCollection 2016.
Meng et al. (2016). Physiological and proteomic responses to salt stress in chloroplasts of diploid and tetraploid black locust (Robinia pseudoacacia L.). Sci Rep. 2016 Mar 15;6:23098. doi: 10.1038/srep23098
Pavlovic et al. (2016). Light-induced gradual activation of photosystem II in dark-grown Norway spruce seedlings. Biochim Biophys Acta. 2016 Feb 18. pii: S0005-2728(16)30028-7. doi: 10.1016/j.bbabio.2016.02.009.
Konert et al.(2015). Protein phosphatase 2A (PP2A) regulatory subunit B'γ interacts with cytoplasmic ACONITASE 3 and modulates the abundance of AOX1A and AOX1D in Arabidopsis thaliana. New Phytol. 2015 Feb;205(3):1250-63. doi: 10.1111/nph.13097. Epub 2014 Oct 13.
Lang et al. (2011).Simultaneous isolation of pure and intact chloroplasts and mitochondria from moss as the basis for sub-cellular proteomics. Plant Cell Rep. 2011 Feb;30(2):205-15.doi: 10.1007/s00299-010-0935-4. - Protocols
-
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extractsPlant Mitochondria Preparation
In this example of mitochondrial preparation used material are etiolated corn seedlings, however, it can also be applied to prepare crude mitochondria from any plant tissue. The volume has to be adjusted, to match the amount of plant material available. For some plant tissues, it can be useful to include sand when grinding the tissue.
Important remarks before you start:
- Whole procedure should be done at 4ºC
- All centrifugations should be done at 4ºC
- Whole procedure should be done as fast as possible
- Plant material should be homogenized very quickly
- Very careful pipeting is recommended
- After isolation membrane integrity can be checked (should be around 80 %). It can be done using Cytochrome C oxidase kit (Sigma)
Materials:
250 ml grinding media (1.8 g PVPP and 0.34 g L-cysteine)
1 liter beaker and large funnel
2 - large, wetted muslins
8 - 35 ml centrifuge tubes
large pestle and mortar
SS-34 rotor Sorvell centrifuges, or JA-20 for Beckman
75 g corn shootsPrecautions:
Until the time of mitochondrial isolation all plants should be kept in the darkness for min. 8 hours. Plant material should not contain any harder plant parts. Proposed tissue amount 15 grams/60 ml of homogenization buffer. Leaf tissue should be quickly and thoroughly cut followed by 3 minute of homogenization.
It is important to keep all materials cold (4°C) and work swiftly while grinding and spinning.
Procedure:
1. Place ½ of the shoots into the mortar and add grinding media to nearly cover the shoots. Grind until shoots are unrecognizable, add additional grinding medium, and pour pulp into muslin and squeeze. Grind the remaining shoots in a similar way and use the second clean muslin to filter. Divide filtrate evenly among the 8 tubes.
2. Spin at 6500 rpm for 2 min at speed.
3. Transfer supernatant into 8 clean tubes.
4. Spin at 12,750 for 5 min at speed.
5. Discard supernatant. Place the tubes on ice at an angle with pellet-side up.
6. Wash the pellets with 1 ml of Wash Media to remove yellow slime
7. Add 5 ml of Wash Media and resuspend the pellet with a pipetman while leaving behind the small white mass (starch). Pool into two tubes.
8. Spin at 6500 rpm for 2 min at speed.
9. Pipet supernatant into clean tubes while leaving behind slimy film
10. Underlay the supernatant with 8 ml of Sucrose cushion.
11. Spin at 9250 rpm for 20 min at speed.
12. Aspirate supernatant.
13. Resuspend each pellet in a suspension medium of choice, this is your crude mitochondrial prearation.Solutions Required
GRINDING MEDIUM: 350 mM Mannitol; 30 mM Mops, 1 mM EDTA, pH 7.6
1 Liter
Mannitol 63.8 g
Mops 6.25 g
1 mM EDTA 2 ml of 0.5 M stock
WASH MEDIUM: 300 mM Mannitol, 20 mM Mops, 1 mM EDTA, pH 7.2250 mls
Mannitol 13.7 g
Mops 1.05 g
1 mM EDTA 0.5 ml of 0.5 M stock
SUCROSE CUSHION: 0.6 M Sucrose100 mls
Sucrose 20.5 g
SUSPENSION MEDIUM: 250 mM Sucrose, 30 mM Mops, pH 7.2100 ml
Sucrose 8.56 g
Mops 0.628 gCourtesy Dr. Thomas Elthon
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extracts
- Reviews:
-
Peter MA | 2025-02-24This antibody works very well for our samples. The total proteins from the Arabidopsis seedlings were subjected to 12% SDS-PAGE and then detected with this antibody (1:4000). The results show that this antibody is quite specific and sensitive for Arabidopsis protein samples.A Podgorska | 2021-05-11sometimes there are two bands for AOX, then it is necessary to add DTT during the extraction of mitochondrial proteins to reduce AOX and get only one band. (Experience with A. thaliana)G. Vanlerberghe | 2018-02-21This antibody worked effectively in Nicotiana tabacum, using either leaf extracts or isolated mitochondria.Michal RUREK | 2014-02-1420 µg of mitochondrial proteins from cauliflower (Brassica oleracea var. botrytis) curds (apical layer) was separated on 12 % SDS-PAGE (Laemmli- type) and blotted 1h to Immobilone P (Millipore) using Sedryt apparatus (Kucharczyk). After blocking (5% Superblock, Pierce) for 1 h at RT, blots were incubated in the primary antibody at a dilution of 1/1000 for overnight at 4 deg. with agitation. Then antibody was additionally bound at 37 deg. for 30 min. The primary antibody, diluted in 5% Superblock solution (Pierce) was used only once. Secondary, anti-rabbit alkaline phosphatase- linked antibodies (ThermoScientific) were bound at 1/10000 dilution at RT (1h). They were diluted in 5% Superblock (Pierce). Three bands of ca. 30-35 kDa were detected using LumiPhos reagent (Pierce). Caution- this antibody does not work with standard ECL system!