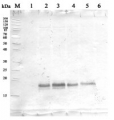
1
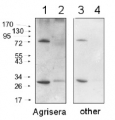
Anti-AtpA | Alpha subunit of ATP synthase, chloroplastic (plant)
AS08 304 | Clonality: Polyclonal | Host: Rabbit | Reactivity: A. thaliana, H. modesticaldum, O. sativa, Z. mays
- Product Info
-
Immunogen: Recombinant maize chloroplast AtpA P05022
Host: Rabbit Clonality: Polyclonal Purity: Serum Format: Lyophilized Quantity: 50 µl Reconstitution: For reconstitution add 50 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Immunogold (IG), Immunoprecipitation (IP), Western blot (WB) Recommended dilution: 1: 200 (IG), 1 : 5 000 (WB) Expected | apparent MW: 55 kDa
- Reactivity
-
Confirmed reactivity: Arabidopsis thaliana, Begonia sp., Capsicum annuum, Heliobacterium modesticaldum, Oryza sativa, Zea mays Predicted reactivity: Dicots and (including Lilium superbum) chloroplast AtpA; may cross-react with mitochondrial AtpA; Algae, Nannochloropsis gaditana, Cyanobacteria
Species of your interest not listed? Contact usNot reactive in: Nicotiana benthamiana - Application Examples
-
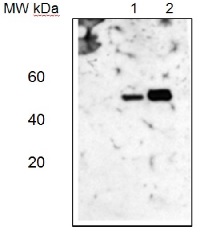
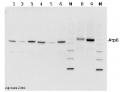
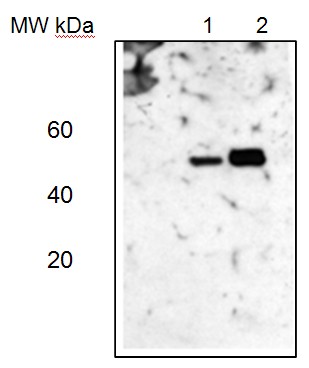 Western blotLane 1: Arabidopsis thaliana total leaf protein extract;Lane 2: Zea mays total leaf protein extract5 µg of total protein were loaded per each lane.Experimental conditions:Proteins were separated in a 12% or 5-15% gradient gel following by a transfer to nitrocellulose membrane. Membrane was blocked with TBST + 4% non-fat dried milk, 20 min following by three washes in TBST. Incubation time with primary and secondary antibodies was 1 hr primary, 30 min for secondary antibodies followed by reaction development using chemiluminescence.
Western blotLane 1: Arabidopsis thaliana total leaf protein extract;Lane 2: Zea mays total leaf protein extract5 µg of total protein were loaded per each lane.Experimental conditions:Proteins were separated in a 12% or 5-15% gradient gel following by a transfer to nitrocellulose membrane. Membrane was blocked with TBST + 4% non-fat dried milk, 20 min following by three washes in TBST. Incubation time with primary and secondary antibodies was 1 hr primary, 30 min for secondary antibodies followed by reaction development using chemiluminescence.Application examples: 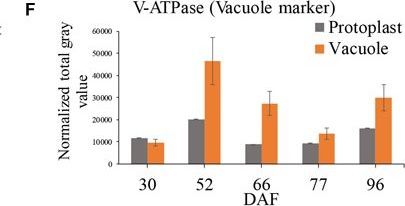
Reactant: Vitis vinifera (Common grape vine)
Application: Western Blotting
Pudmed ID: 31156689
Journal: Front Plant Sci
Figure Number: 2F
Published Date: 2019-06-04
First Author: Kuang, L., Chen, S., et al.
Impact Factor: 5.435
Open PublicationGrape berry flesh cell protoplast and vacuole preparations and quality check. (A) Isolated protoplasts. (B) Vacuole enrichment by Ficoll gradient centrifugation. (C) Crude vacuoles stained with Neutral Red. (D) Vacuoles in vacuole-enriched preparation stained with Neutral Red, solid arrow indicating vacuole, dashed arrow indicating tonoplast vesicle, inset indicating vacuoles with microdomains. (E) Relative quantity of P-ATPase between protoplast and vacuole isolations by western blotting analysis. (F) Relative quantity of V-ATPase between protoplast and vacuole isolations by western blotting analysis. Values were the average of three replicates ± SD.
- Additional Information
-
Additional information: Sequence of the protein used for eliciting this antibody is also conserved in Arabidopsis thaliana AtpA P56757
- Background
-
Background: ATP synthase is the universal enzyme that synthesizes ATP from ADP and phosphate using the energy stored in a transmembrane ion gradient. AtpA is the largest subunit of the membrane-extrinsic ATP synthase subcomplex.
- Product Citations
-
Selected references: Uflewski et al. (2024). The thylakoid proton antiporter KEA3 regulates photosynthesis in response to the chloroplast energy status. Nat Commun. 2024 Mar 30;15(1):2792. doi: 10.1038/s41467-024-47151-5.
Chen et al. (2023) Variation of CRS2 Affected the Establishment of Photosynthetic System in Rice. Int J Mol Sci. 2023 Mar 18;24(6):5796. doi: 10.3390/ijms24065796.
Harchouni et al. (2022) Guanosine tetraphosphate (ppGpp) accumulation inhibits chloroplast gene expression and promotes super grana formation in the moss Physcomitrium (Physcomitrella) patens. New Phytol. 2022;236(1):86-98. doi:10.1111/nph.18320.
Kuang et al. (2019). Quantitative Proteome Analysis Reveals Changes in the Protein Landscape During Grape Berry Development With a Focus on Vacuolar Transport Proteins. Front Plant Sci. 2019 May 15;10:641. doi: 10.3389/fpls.2019.00641. eCollection 2019.
Pao et al. (2018). Lamelloplasts and minichloroplasts in Begoniaceae: iridescence and photosynthetic functioning. J Plant Res. 2018 Mar 2. doi: 10.1007/s10265-018-1020-2. (ImmunoGold)
Zhang et al. (2017). Nitric oxide induces monosaccharide accumulation through enzyme S-nitrosylation. Plant Cell Environ. 2017 Sep;40(9):1834-1848. doi: 10.1111/pce.12989.
Jeon et al. (2017). Functional characterization of chloroplast-targeted RbgA GTPase in higher plants. Plant Mol Biol. 2017 Nov;95(4-5):463-479. doi: 10.1007/s11103-017-0664-y.
Murcha et al. (2016). Plant specific Preprotein and Amino Acid Transporter proteins are required for tRNA import into mitochondria. Plant Physiol. 2016 Oct 27. pii: pp.01519.2016.
Camejo et al. (2015). Proteomic identification of mitochondrial carbonylated proteins in two maturation stages of pepper fruits. Proteomics. 2015 Aug;15(15):2634-42. doi: 10.1002/pmic.201400370.
Yang et al. (2015). Purification and biochemical characterization of the ATP synthase from Heliobacterium modesticaldum. Protein Expr Purif. 2015 May 12. pii: S1046-5928(15)00111-4. doi: 10.1016/j.pep.2015.05.006. - Protocols
-
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extracts
Oxygenic photosynthesis poster by prof. Govindjee and Dr. Shevela
Z-scheme of photosynthetic electron transport by prof. Govindjee and Dr. Björn and Dr. ShevelaExtraction of Total Leaf Proteins
• KEEP EVERYTHING COLD AT ALL TIMES
• DO NOT ALLOW LEAF SAMPLES TO COME CLOSE TO THAWING UNTIL GRINDING IN HOMOGENIZATION BUFFER.
• KEEP EXTRACTED SAMPLES OUT OF THE –80 ˚ FREEZER FOR THE MINIMAL AMOUNT OF TIME TO AVOID PROTEIN DEGRADATION. Remove samples from freezer immediately prior to use (i.e. after tubes with SDS-sample buffer are labelled and prepared) and return immediately after pipetting out aliquots for analysis. If extracting many samples, extract in batches of six, spot onto Coomassie for quantification (see below) and place in –80 freezer before extracting the next batch. When experienced, it should be possible to do 18 preps in 30 minutes. When starting out, plan to do 12 preps in 30 minutes, in two groups of six.
• Protein Quantification: If care is taken when harvesting tissue to always harvest the same amount (e.g. a 1 cm leaf tip of the 2nd leaf), there is no need to quantify protein by Coomassie spotting. Otherwise, use the simple quantification procedure outlined below.
• Extract protein from several wild-type seedling that were grown in parallel with each batch of mutants. The same wild-type controls can be used for all plants that were planted and harvested at the same time and place.
1. Make a list of samples that will be extracted. Label 1.4 ml eppendorf tubes with fine-tipped black sharpie (top: seedling number and phenotype (e.g. F235-7-2 pg) , side: seedling number, date (month/year). It is important to number on top and side because numbers can rub off.
Gather materials: mortars and pestles (6 small ones), liquid nitrogen (if working with frozen samples), protein extraction buffer (see below), gray eppendorf tube rack with ice, Whatman paper for spotting to quantify protein; label with pencil the positions where each sample will be spotted.
2. Prepare homogenization buffer. Add the labile additives described below (ß-ME, PMSF, leupeptin and pepstatin) to a 5 ml aliquot of the Homogenization Buffer Base Stock. This will be sufficient to prep ~ 20 samples. Store the complete buffer on ice and discard unused buffer after 30 minutes.
Protein Homogenization Buffer Base Stock (store at 4°):
For 100 ml: 10 ml 1M Tris-HCl pH 7.5
20 ml 50% glycerol
1 ml 0.5 M EDTA
2 ml 0.1M EGTA
67 ml ddH20
Before use, aliquot sufficient buffer for 30 minutes of extractions (e.g. 5 ml) and add:
* PMSF to 2 mM (1/20th volume of a 40 mM stock in isoproponol stored at –20 degrees. The stock should be mixed and heated sufficiently to put any crystals into solution before pipetting. PMSF is toxic and inactivates within 30 minutes after adding to aqueous solution).
* ß-mercaptoethanol (ß-ME) to 40 mM. (27µl of the pure liquid stock (stored in hood) per 10 ml)
* Pepstatin to 2 µg/ml. (1/500th volume of a 1 mg/ml stock. The stock is stored at –20˚.)
* Leupeptin to 2 µg/ml (1/5000th volume of a 10 mg/ml stock. The stock is stored at –20˚)
For 5 mls of complete buffer add: 13 µl B-ME, 10 µl 1 mg/ml pepstatin, 250 µl 40 mM PMSF and 1 µl 10 mg/ml leupeptin. Discard this buffer after 30 minutes.
3. Grind tissue. Grind in small mortar and pestle.
If using fresh tissue, it is simplest to grind the unfrozen tissue directly in ice cold homogenization buffer (200 µl) in pre-chilled mortar and pestle.
If using frozen tissue, grind tissue to a fine powder in liquid nitrogen and then add 200 µl homogenization buffer to the frozen powder and continue grinding to a paste. It is essential to keep the tissue frozen until the buffer is added.
Must grind hard to break bundle sheath cells. Failure to do this will reduce the yield of Rubisco and other bundle sheath- specific proteins.
4. For Total Leaf Extract (the usual case for routine mutant screening), pipet the extract into the labelled eppendorf tube, on ice. If quantifying by Coomassie spotting (i.e. if the amount of tissue harvested wasn’t similar for all samples), spot 2 µl onto Whatman 3MM paper (label spot with pencil).
For analysis of separated membrane and soluble proteins: pellet the membranes by microfuging for 3 minutes at top speed in the microfuge in the cold; Pipet off the sup avoiding the green pellet, and place in a new microfuge tube. This is the "soluble" fraction. Rinse the membranes to be sure they are free of soluble proteins: Add 0.2 ml homogenization buffer to each membrane sample, flick gently with your finger, and microfuge for 2 minutes. Discard the sup; Resuspend the membrane pellets in 50-200 µl homogenization buffer, by pipeting up and down gently. For a series of samples that you want to compare on a gel, resuspend the smallest green pellet in 50 µl, and the remaining pellets in whatever volume it takes to give an equal density of greenness (to your eye).
5. Store the protein samples in the -80o freezer. Repeated freeze-thaw cycles can lead to protein degradation. So for particularly precious samples, freeze them in small aliquots.
6. Quantify the protein. If samples contained uneven amounts of leaf tissue, the relative protein concentrations can be estimated by the following simple procedure.
Spot 2 µl of each sample on toWhatman 3mm (with samples labelled).
To aid in quantification, spot 1:2 dilutions of several samples that you expect are particularly concentrated (i.e. they are particularly green). Each spot should have the same volume- i.e. on piece of parafilm, mix 2 µl of sample to 2 µl water, and then spot 2 µl of this onto the Whatman paper. This will allow you to see what 1/2 as much protein looks like. For even better quantification, spot 1,3, and 10 ug of BSA in a 2 ul volume.
Place the Whatman 3MM paper with the protein spots in a container that has previously been used for Coomassie staining (i.e. it is stained blue). Add the Coomassie staining solution (325 ml water, 125 ml isoproponol, 50 ml acetic acid, 250 mg Coomassie Blue R-250). Rock briefly until you can see the spots turn blue (10-20 seconds). Rinse with water until the background is fairly white. Compare signal intensities while paper is either fully wet or fully dry. If it is partially dry, results may be misleading.
Gels- flick tube to suspend stuff. Pipet out what you want put in sample, heat to 70ish, 1 min spin
Courtesy Dr. Alice Barkan Lab, University of Oregon, USA - Reviews:
-
This product doesn't have any reviews.
Accessories
AS05 085 | Clonality: Polyclonal | Host: Rabbit | Reactivity: [global antibody] for plant, green alga, animal and bacterial F-type ATP synthases
Benefits of using this antibody