1
Anti-AtpB | Beta subunit of ATP synthase (chloroplastic + mitochondrial) (rabbit antibodies)
AS05 085 | Clonality: Polyclonal | Host: Rabbit | Reactivity: [global antibody] for plant, green alga, animal and bacterial F-type ATP synthases
Benefits of using this antibody
- Product Info
-
Immunogen: KLH-conjugated synthetic peptide derived from available plant, algal (chloroplastic and mitochondrial) and bacterial sequences of beta subunits of F-type ATP synthases, including Arabidopsis thaliana chloroplastic ATP synthase subunit beta UniProt: P19366, TAIR: AtCg00480 and Arabidopsis thaliana mitochondrial ATP synthase subunit beta-1, UniProt: P83483, TAIR: At5g08670 as well as Chlamydomonas reinhardtii, UniProt: P06541 and A8IQU3
Host: Rabbit Clonality: Polyclonal Purity: Serum Format: Lyophilized Quantity: 50 µl Reconstitution: For reconstitution add 50 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Blue Native-PAGE (BN-PAGE), Immunofluorescence (IF), Ultrastructure Expansion Microscopy (U-ExM), Western blot (WB) Recommended dilution: 1 : 100 (IF), 1 : 5000 (BN-PAGE), 1: 2500 (U-ExM), 1 : 2000-1 : 5 000 (WB) Expected | apparent MW: 53.9 kDa (Arabidopsis thaliana), 51.7 kDa (Synechocystis PCC 6803), 53.7 kDa (Spinacia oleracea)
- Reactivity
-
Confirmed reactivity: Arabidopsis thaliana, Bacillus cereus, Bryopsis corticulans, Camelina sinensis, Chlamydomonas reinhardtii, Chlorella vulgaris, Chromochloris zofingiensis, Cyanidioschyzon merolae, Dionaea muscipula, Echinochloa crus/galli, Escherichia coli, Gossypium hirsutum, Helicobacter pylori, Hordeum vulgare, Gladieria sulphuraria, Glycine max, Lycopersicum esulentum, Moniliophthora perniciosa, Nannochloropsis salina, Neochloris oleoabundans (chlorophyta), Nicotiana bentamiana, Nicotiana tabacum, Oryza sp. (roots, leafs, pollen), Ostreococcus tauri, Pheodactylum tricornutum CCAP 1055/1, Pisum sativum, Plasmodium berghei, Populus sp., Robinia pseudoacacia, Scaphoideus titanus, Selaginella martensii, Setaria viridis, Solanum lycopersicum, Spinacia oleracea, Tetrabaena socialis, Toxoplasma gondii, Zea mays
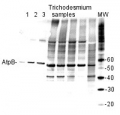
Animal tissues from: cow, chicken, pig, rat, salmon, seal, Locusta migratoriaPredicted reactivity: Acinetobacter baumannii, Algae, Bacillus subtilis, Brassica napus, Cannabis sativa, Clostridioides difficile, Cyanobacteria, E.coli K-12, Eragrostis tef, Galdieria sulphuraria, Heliomicrobium modesticaldum Ice1, Lolium multiflorum, Manihot esculenta, Nicotiana plumbaginifolia, Saccharomyces cerevisiae, Salmonella typhimurium, Trichodesmium erythraeum, Triticum aestivum, Vitis vinifera, Zosteria marina, Yrsinia sp.
Species of your interest not listed? Contact usNot reactive in: Archeal V-type ATP synthase - Application Examples
-
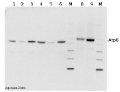
2 µg of total protein extracted with PEB (AS08 300) from leaf tissue of (1) Arabidopsis thaliana, (2) Spinacia oleracea, (3) Lycopersicon esculentum, (4) Glycine max, (5) Populus sp., (6) Zea mays and (7) Hordeum vulgare were separated on 4-12% NuPage (Invitrogen) LDS-PAGE and blotted 1h to nitrocellulose. In parallel a dilution row (a-g: 10 - 5 - 2.5 - 1.25 - 0.63 - 0.32 - 0.16 µg protein/lane) from sample 1 (Arabidopsis) was processed. Filters were blocked 1h with 2% low-fat milk powder in TBS-T (0.1% TWEEN 20) and probed with anti-AtpB (AS08 085, 1:5000, 1h) and secondary anti-rabbit (1:10000, 1 h) antibody (HRP conjugated, recommended secondary antibody AS09 602) in TBS-T containing 2% low fat milk powder. Antibody incubations were followed by washings in TBS-T (15, +5, +5, +5 min). All steps were performed at RT with agitation. Signal was detected with chemiluminescent substrate, using a Fuji LAS-3000 CCD (300s, standard sensitivity).
.jpg)
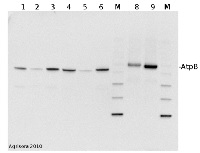
2 µg of total protein from (1) cow muscle, (2) chicken muscle, (3) pig muscle, (4) rat liver, (5) salmon muscle, (6) seal muscle, (8) Arabidopsis thaliana, (9) Zea mays extracted with Protein Extration Buffer, PEB (AS08 300) and separated on 4-12% NuPage (Invitrogen) LDS-PAGE and blotted 1h to PVDF. Blots were blocked immediately following transfer in 2% blocking reagent in 20 mM Tris, 137 mM sodium chloride pH 7.6 with 0.1% (v/v) Tween-20 (TBS-T) for 1h at room temperature with agitation. Blots were incubated in the primary antibody at a dilution of 1: 50 000 for 1h at room temperature with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, then washed once for 15 min and 3 times for 5 min in TBS-T at room temperature with agitation. Blots were incubated in secondary antibody (Agrisera anti-rabbit IgG horse radish peroxidase conjugated, AS09 602) diluted to 1:50 000 in 2% blocking solution for 1h at room temperature with agitation. The blots were washed as above and developed for 5 min with chemiluminescent detection reagent according to the manufacturers instructions. Images of the blots were obtained using a CCD imager (FluorSMax, Bio-Rad) and Quantity One software (Bio-Rad). Exposure time was 30 seconds.
M - molecular weight marker
Application examples: 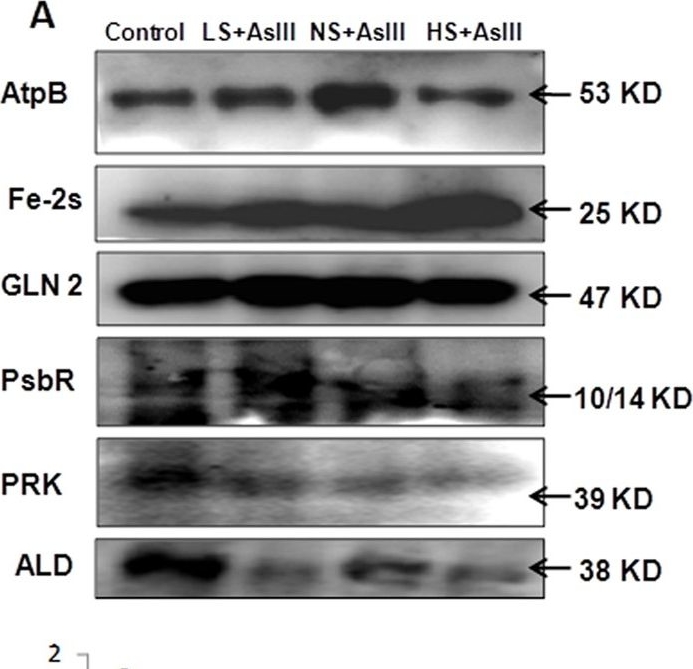
Reactant: Oryza sativa (Asian rice)
Application: Western Blotting
Pudmed ID: 26552588
Journal: Sci Rep
Figure Number: 6A
Published Date: 2015-11-10
First Author: Dixit, G., Singh, A. P., et al.
Impact Factor: 4.13
Open PublicationWestern blot analysis of selected candidate proteins (AtpB, 2Fe-2S, GLN2, Psb R, PRK, ALD) with their corresponding molecular weight in rice leaves during As stress under various S regimes (A); RuBisCo large subunit a band in coomassie blue staining (CBB) SDS gel served as a loading control (B), which used for normalization of detected proteins in densitometry studies (C) fold change indicated with respect of control samples.
Reactant: Nicotiana tabacum (Common tobacco)
Application: Western Blotting
Pudmed ID: 28180288
Journal: J Exp Bot
Figure Number: 5A
Published Date: 2017-02-01
First Author: Schöttler, M. A., Thiele, W., et al.
Impact Factor: 6.088
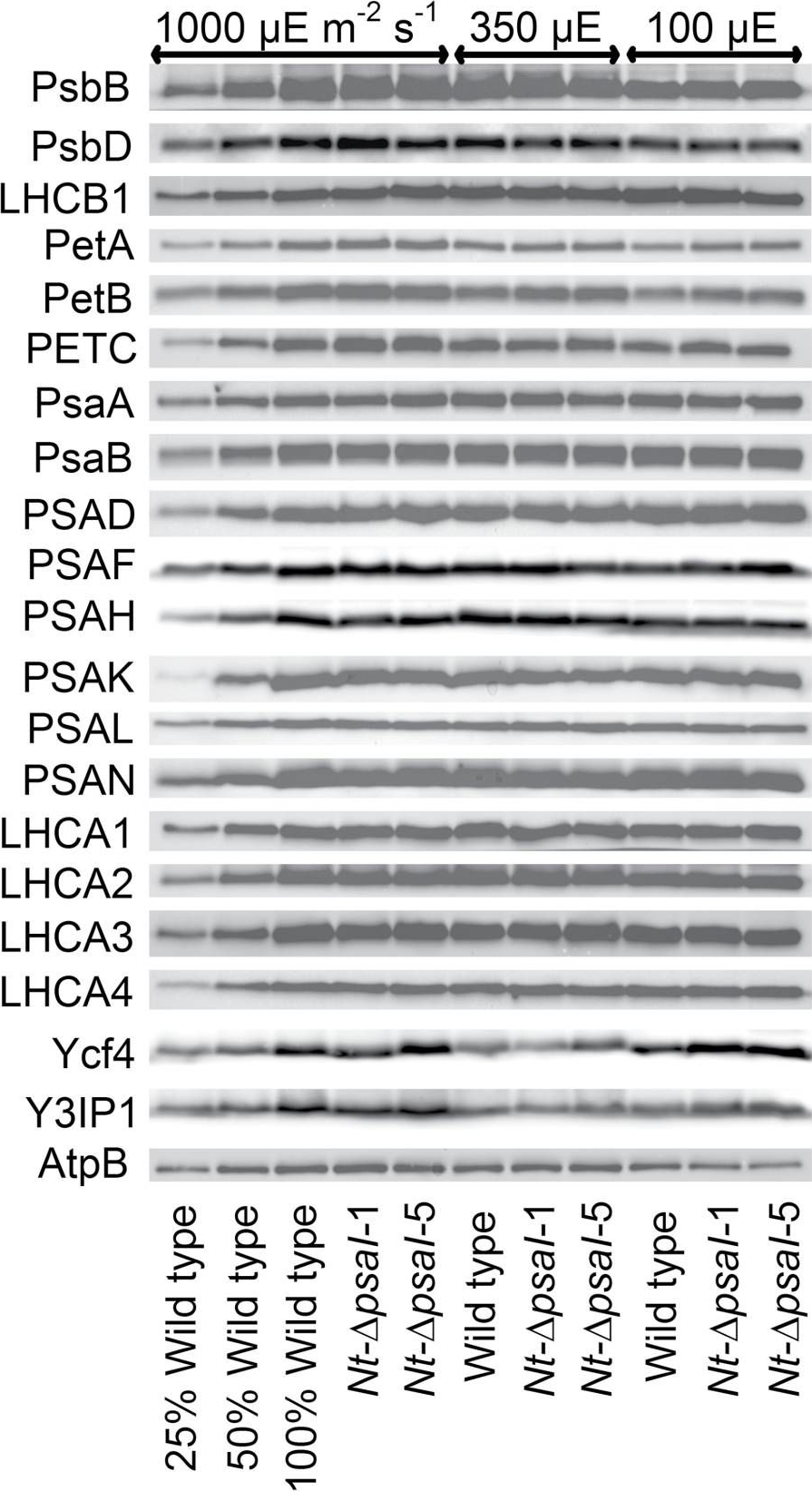
Open PublicationImmunoblot analysis of photosynthetic complex accumulation in wild-type tobacco and the two ?psaI lines grown under low, intermediate, and high-light conditions. Because the accumulation of most tested proteins was highest under high-light conditions, lanes one to three contain samples diluted to 25%, 50%, and a 100% sample of wild-type tobacco grown under high-light conditions, to allow for semi-quantitative determination of changes in protein abundance. Lanes four and five contain the two transplastomic lines grown at 1000 µE m?2 s?1. Lanes six to eight contain wild-type tobacco and the mutants grown at intermediate light intensities, and lanes nine to eleven contain samples grown at low light intensities. For PSII, the accumulation of the essential subunits PsbB (CP43) and PsbD (D2) and the LHCB1 antenna protein were determined, while for the cytochrome b6f complex, the accumulation of the essential redox-active subunits PetA (cytochrome f), PetB (cytochrome b6), and PETC (Rieske FeS protein) was tested. AtpB was probed as an essential subunit of the chloroplast ATP. For PSI, in addition to the three essential plastome-encoded subunits PsaA, PsaB, and PsaC, the accumulation of the nuclear-encoded subunits PSAD, PSAH, PSAK, PSAL, and PSAN and of the four LHCI proteins (LHCA1, LHCA2, LHCA3, LHCA4) was determined. Finally, we examined the accumulation of Ycf4, the chloroplast-encoded PSI-biogenesis factor encoded in the same operon as PsaI, and the nuclear-encoded assembly factor Y3IP1.
Reactant: Algae
Application: Western Blotting
Pudmed ID: 29460179
Journal: Planta
Figure Number: 7A,B,C
Published Date: 2018-06-01
First Author: Giovagnetti, V., Han, G., et al.
Impact Factor: 3.95
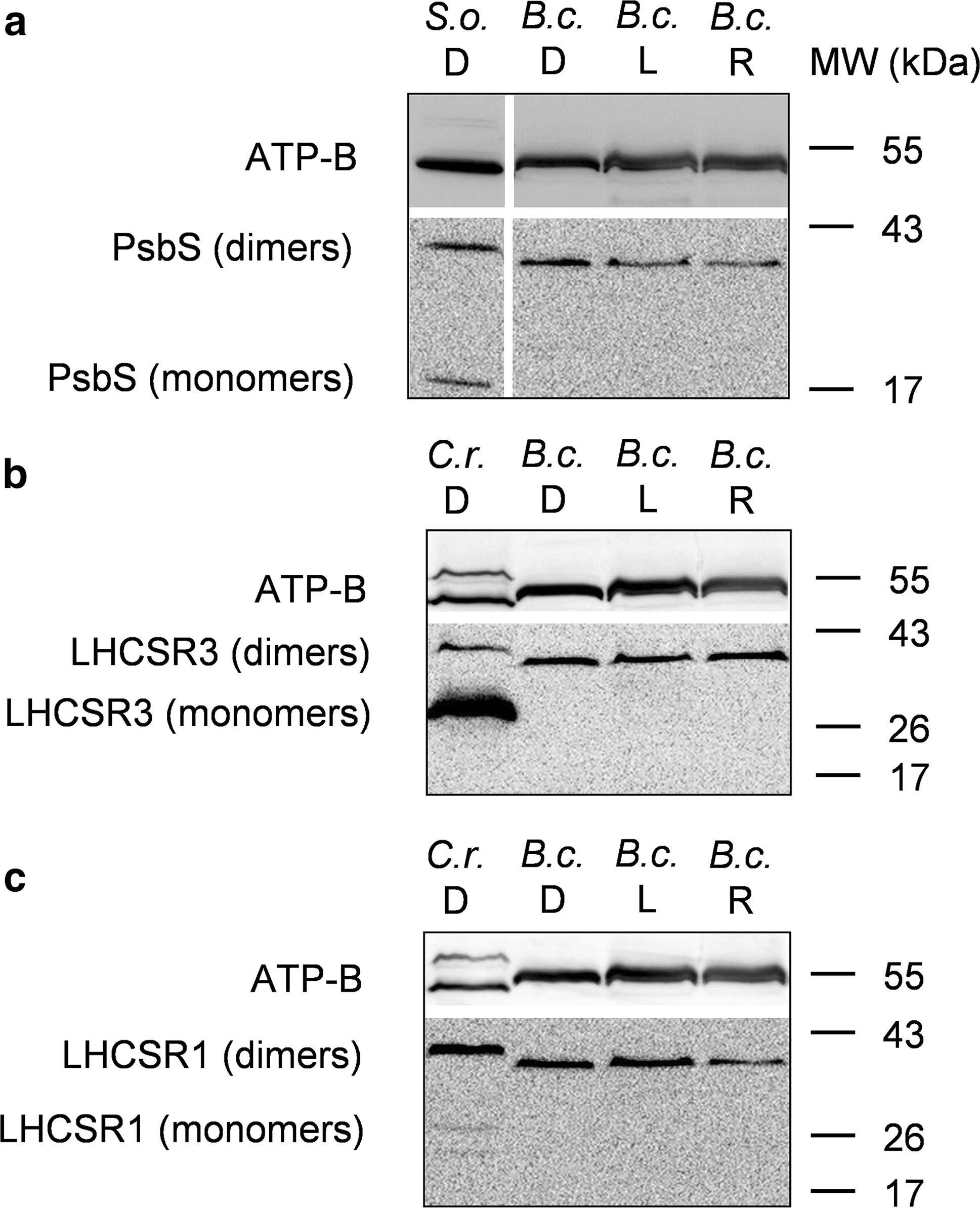
Open PublicationWestern blot analysis of PsbS and LHCSR proteins from dark-adapted (D, 30 min), light-treated (L, 2500 µmol photons m?2 s?1, 30 min) and post-dark-recovery chloroplasts (R, 1 h) isolated from B. corticulans. Samples were analysed by immunoblotting with an anti-PsbS (a), anti-LHCSR3 (b) and anti-LHCSR1 antibodies (c). Control samples are dark-adapted S. oleracea (a) and C. reinhardtii chloroplasts (b, c). Note that B. corticulans and S. oleracea samples have been probed against anti-PsbS antibody in the same membrane. The separation between lanes is only due to the presence of different samples between those presented of S. oleracea and B. corticulans (a). The ? subunit of ATP synthase (ATP-B) was used as loading control. 10 µg of Chl was loaded in each lane
Reactant: Chlamydomonas reinhardtii (Green Alga)
Application: Western Blotting
Pudmed ID: 30808958
Journal: Sci Rep
Figure Number: 2B
Published Date: 2019-02-26
First Author: Tokutsu, R., Fujimura-Kamada, K., et al.
Impact Factor: 4.13
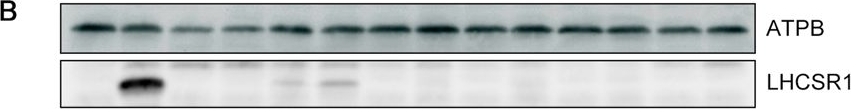
Open PublicationLHCSR1 expression in isolated mutants following UV induction. (A) Gene expression analysis of LHCSR1 by RT-PCR. The CBLP gene was used as the housekeeping control. RNA and protein samples were collected after 1?hour of irradiation with UV. A 100-bp DNA ladder is shown. (B) Expression of the LHCSR1 protein was detected using specific antibodies, and the ATPB protein was used as the loading control. The protein samples were collected after 4?hours of irradiation with UV. Samples illustrated here are representative of three biological replicates.
Reactant: Chlamydomonas reinhardtii (Green Alga)
Application: Western Blotting
Pudmed ID: 31506429
Journal: Nat Commun
Figure Number: 1E
Published Date: 2019-09-10
First Author: Tokutsu, R., Fujimura-Kamada, K., et al.
Impact Factor: 13.783
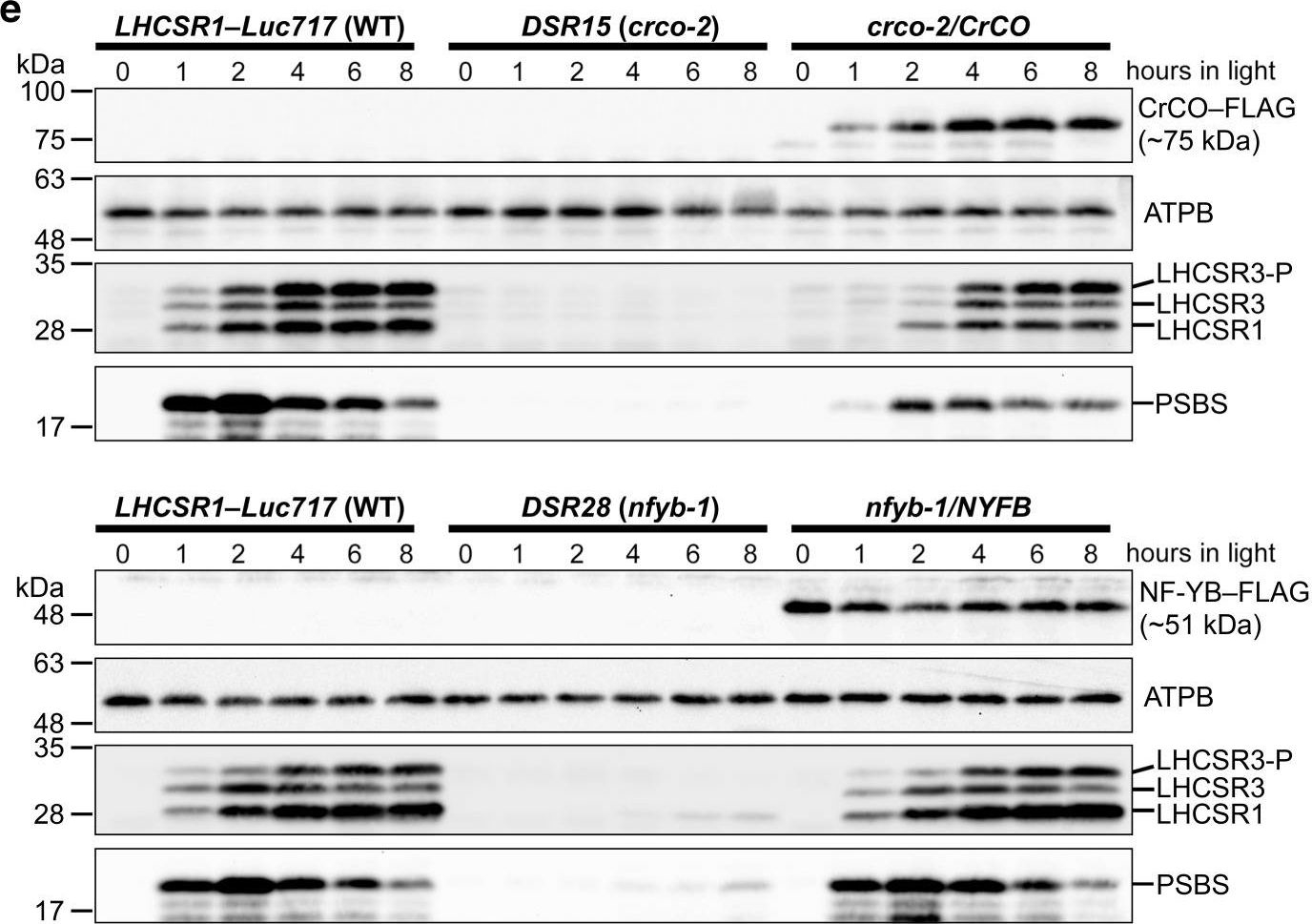
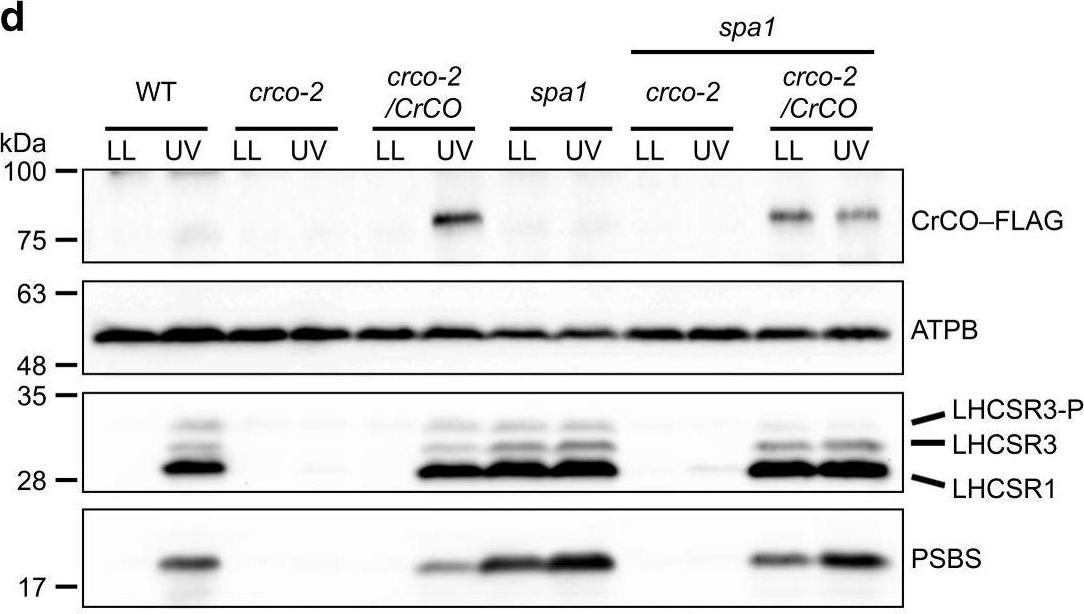
Open PublicationCrCO and NF-YB are crucial for photoprotection. a The bleaching phenotypes of the reference strain (LHCSR1–Luc717) and the DSR mutants visualized in multiwell plates. Representative cell cultures treated with low light (LL; left wells) or high light (HL; right wells). Concentrations of the cultures were adjusted to 1.0?×? 107 cells/mL. b Chlorophyll content per cell after LL (closed bar) or HL (open bar) treatment of the cells shown in a. c Maximum quantum yield of photosystem II (Fv/Fm) during HL treatment. d qE quenching capability during HL treatment. e Immunoblot analysis of 3xFLAG-fused proteins (CrCO–FLAG and NF-YB–FLAG in crco-2/CrCO and nfyb-1/NFYB, respectively), LHCSR1, LHCSR3, and PSBS during HL treatment. ATPB protein levels are shown as the loading control. The experiments were performed three times with different biological samples (n?=?3 biological replicates; mean?± S.E.M); a representative experiment is shown in e
Reactant: Chlamydomonas reinhardtii (Green Alga)
Application: Western Blotting
Pudmed ID: 31506429
Journal: Nat Commun
Figure Number: 3D
Published Date: 2019-09-10
First Author: Tokutsu, R., Fujimura-Kamada, K., et al.
Impact Factor: 13.783
Open PublicationUVR8 interacts with COP1 and SPA1 to activate the CrCO-based transcriptional complex under UV irradiation. a Confocal images of UVR8–Venus–3xFLAG proteins in the DSR1–comp15 (uvr8/UVR8) strain. The cells were treated with UV for 30?min. Scale bars, 10?µm. b After 1?h of UV treatment, the cells were harvested and UVR8–Venus–3xFLAG proteins were immunoprecipitated by FLAG (M2) antibody with SURE-beads. The coimmunoprecipitated proteins were identified by LC-MS/MS analysis of the Coomassie Brilliant Blue-stained polypeptide bands obtained via SDS-PAGE separation after in-gel trypsin digestion. M, molecular mass standard. c Interaction profiles among COP1, SPA1, and CrCO visualized with Y2H assays, which were performed using COP1, SPA1, and CrCO fused with the AD and/or BD domains of GAL4. The culture conditions were as described for Fig. 2b. d Immunoblot analysis of LHCSRs, PSBS, and 3xFLAG-fused CrCO to visualize protein accumulation in the wild-type control (WT), crco-2, crco-2/CrCO, spa1, spa1 crco-2, and spa1 crco-2/CrCO strains before and after UV treatment. ATPB proteins were included as the loading controls
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 33322466
Journal: Biomolecules
Figure Number: 7A
Published Date: 2020-12-11
First Author: Andreeva, A. A., Vankova, R., et al.
Impact Factor: None
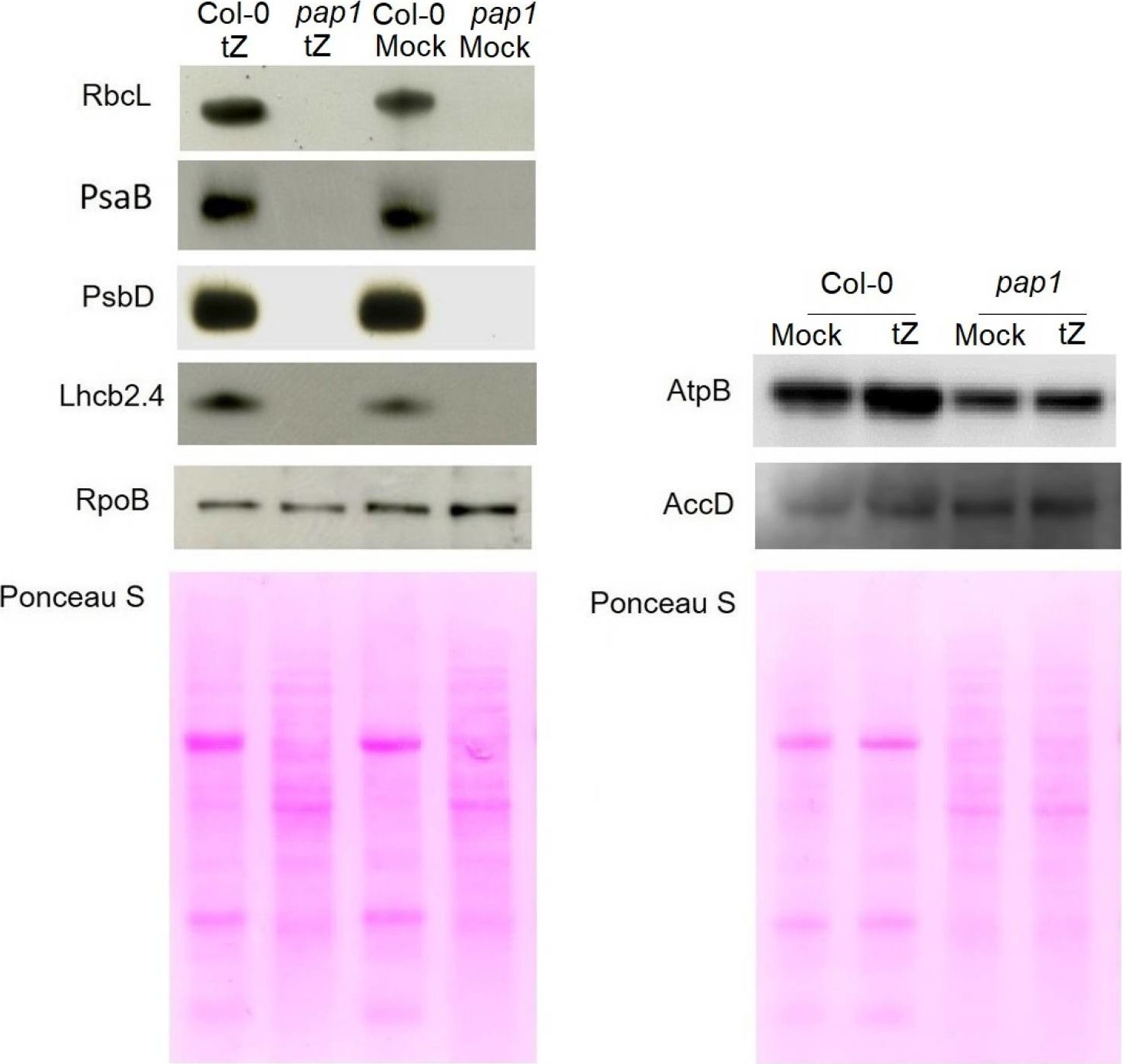
Open PublicationImmunoblot analysis of the photosynthetic proteins on the basis of equal total Ponceau S dye stained blot with proteins from leaves of wild type plants and pap1 mutant grown on MS medium in Petri dishes for four weeks under a 16 h light/8 h dark photoperiod at 23 °C with 100 ?E m?2 s?1. Proteins were visualized by immunoblotting using antibodies specific for RbcL, PsaB, PsbD, AtpB, RpoB, AccD and Lhcb2.4 proteins.
Reactant: Solanum lycopersicum (Tomato)
Application: Western Blotting
Pudmed ID: 34439953
Journal: Biology (Basel)
Figure Number: 9A
Published Date: 2021-07-28
First Author: Trojak, M. & Skowron, E.
Impact Factor: None
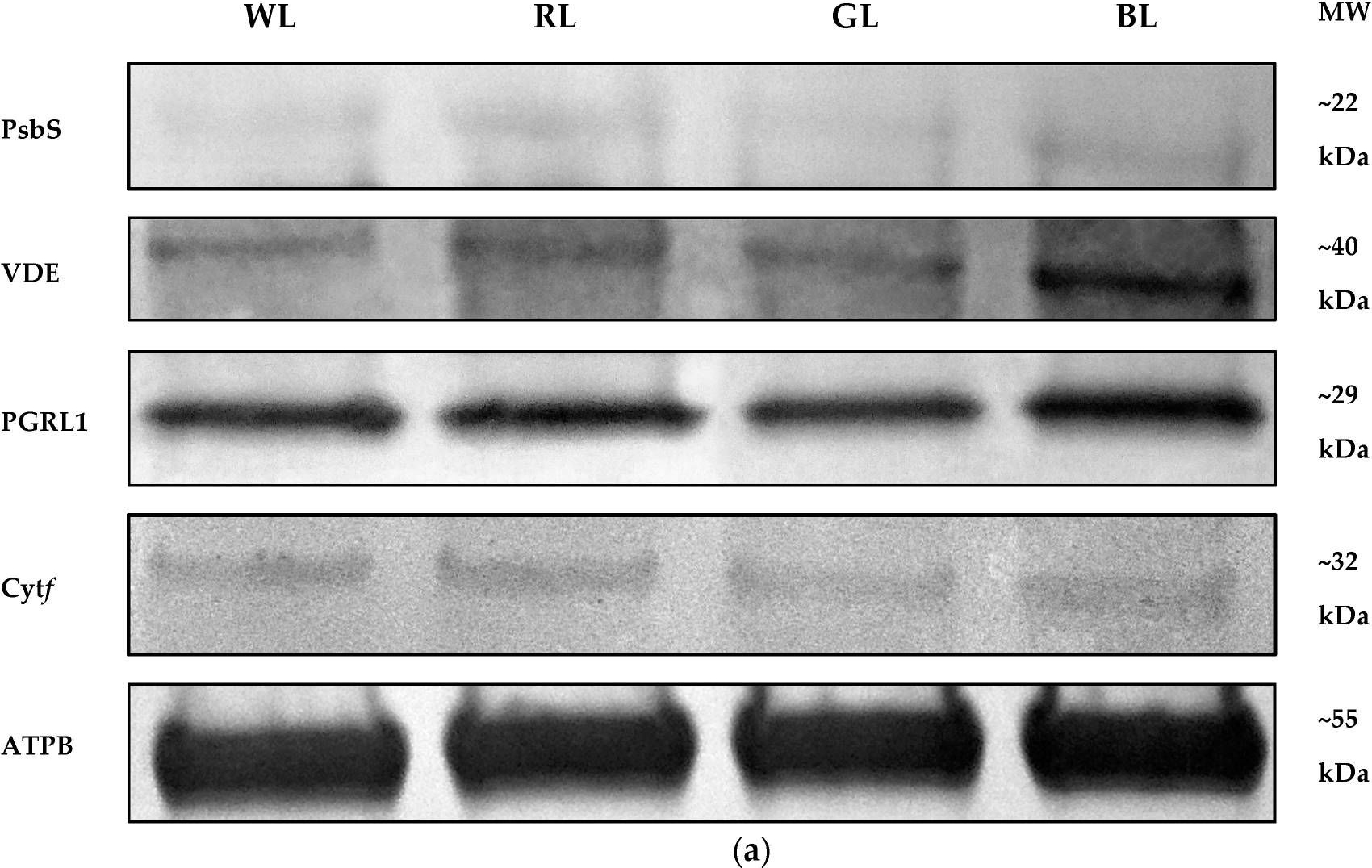
Open PublicationWestern Blot analyses (a) and densitometric analyses of PsbS (b), VDE (c), PGRL1 (d), and cytf (e) proteins in leaves of tomato plants (Solanum lycopersicum L. cv. Malinowy Ozarowski) grown under different light conditions (see Material and Methods for details). The bands were normalized to the appropriate ? subunit of ATP synthase (ATPB) band (loading control, Figure S4) (a). The bar diagrams (b–e) represent pixel volumes (densitometric analyses) of proteins in samples. Each value represents the mean ± SD (n = 3) considering the control sample (WL) value as 1 (100%). Different letters indicate significant differences between treatments (p = 0.05) with a Tukey’s HSD test. MW—molecular weight.
- Additional Information
-
Additional information: The anti-AtpB antibody will detect the mitochondrial form of the F1 ATP synthase subcomplex, as well as the chloroplastic CF1 Atp Synthase, and most known bacterial F-type Atp Synthases. Peptide used for antibody production is located in a beta sheet, which is partly exposed near the surface of the AtpB protein.
Anti-AtpB antibody was used as a loading control in Chlamydomonas reinhardtii and Synechocystis sp. PCC6803.
This product can be sold containing proclin if requestedAdditional information (application): Blue Native gel electrophoresis (BN-PAGE) has been performed on samples solubilized with digitonin (4:1) and loaded at 100 µg/well. Gel thickness was 2 mm with 4.5-16 % gradient.Antibody is recognizing mitochondrial form of AtpB Subota el. al (2011).
This antibody can be used as a loading control for bacteria, Bacillus cereus. - Background
-
Background: ATP synthase is the universal enzyme that synthesizes ATP from ADP and phosphate using the energy stored in a transmembrane ion gradient.
- Product Citations
-
Selected references: Mohamed et al. (2026). Unraveling design principles of protein landscapes in photosynthetic membranes in plant chloroplasts. Sci Adv. 2026 Feb 13;12(7):eaeb2410. doi: 10.1126/sciadv.aeb2410.
Lv et al. (2025). Plant-derived hydrogel and photosynthetic nano-units for myocardial infarction therapy.Nat Commun. 2025 Aug 18;16(1):7678. doi: 10.1038/s41467-025-62020-5.
Wójtowicz et al. (2025). Shrink or expand? Just relax! Bidirectional grana structural dynamics as early light-induced regulator of photosynthesis. New Phytol . 2025 Jun;246(6):2580-2596. doi: 10.1111/nph.70175.
Shikha et al. (2025). Numerous rRNA molecules form the apicomplexan mitoribosome via repurposed protein and RNA elements. Nat Commun. 2025 Jan 18;16(1):817. doi: 10.1038/s41467-025-56057-9.
Maclean et al. (2024). The Toxoplasma gondii mitochondrial transporter ABCB7L is essential for the biogenesis of cytosolic and nuclear iron-sulfur cluster proteins and cytosolic translation. mBio. 2024 Oct 16;15(10):e0087224. doi: 10.1128/mbio.00872-24.
Kubota et al. (2024). The blue–green light-dependent state transition in the marine phytoplankton Ostreococcus tauri. New Phytol. 2024 Sep 23. doi: 10.1111/nph.20137
Ermakova et al. (2024). Chloroplast NADH dehydrogenase-like complex-mediated cyclic electron flow is the main electron transport route in C4 bundle sheath cells. New Phytol. 2024 Jul 22.doi: 10.1111/nph.19982.
Hoa et al. (2024). Proteomic analysis on symbiotic differentiation of mitochondria in soybean nodules. Comparative Study Plant Cell Physiol. 2004 Mar;45(3):300-8. doi: 10.1093/pcp/pch035.
Zhao et al. (2024). Psb28 protein is indispensable for stable accumulation of PSII core complexes in Arabidopsis.Plant J. 2024 May 26. doi: 10.1111/tpj.16844.
Ciesielska et al. (2024). S2P2-the chloroplast-located intramembrane protease and its impact on the stoichiometry and functioning of the photosynthetic apparatus of A. thaliana. Front Plant Sci. 2024 Mar 15:15:1372318. doi: 10.3389/fpls.2024.1372318.
Usey and Huet et al. (2023). ATP synthase-associated coiled-coil-helix-coiled-coil-helix (CHCH) domain-containing proteins are critical for mitochondrial function in Toxoplasma gondii. mBio. 2023 Oct 31;14(5):e0176923. doi: 10.1128/mbio.01769-23.
Sulli et al. (2023). Generation and physiological characterization of genome‑edited Nicotiana benthamiana plants containing zeaxanthin as the only leaf xanthophyll. Planta . 2023 Oct 5;258(5):93. doi: 10.1007/s00425-023-04248-3.
Jiang et al. (2023). Toxic effects of lanthanum (III) on photosynthetic performance of rice seedlings: Combined chlorophyll fluorescence, chloroplast structure and thylakoid membrane protein assessment. Ecotoxicol Environ Saf. 2023 Nov 15:267:115627.doi: 10.1016/j.ecoenv.2023.115627.
Kafri et al. (2023). Systematic identification and characterization of genes in the regulation and biogenesis of photosynthetic machinery. Cell. 2023 Dec 7;186(25):5638-5655.e25.doi: 10.1016/j.cell.2023.11.007.
Silva et al.(2023). Functional and biochemical characterisation of the Toxoplasma gondii succinate dehydrogenase complex.PLoS Pathog. 2023 Dec 11;19(12):e1011867.doi: 10.1371/journal.ppat.1011867.
Ye et al. (2023). The light-harvesting chlorophyll a/b-binding proteins of photosystem II family members are responsible for temperature sensitivity and leaf color phenotype in albino tea plant. J Adv Res . 2023 Dec 25:S2090-1232(23)00404-6.doi: 10.1016/j.jare.2023.12.017.
Hao and Malnoë (2023). A Simple Sonication Method to Isolate the Chloroplast Lumen in Arabidopsis thaliana.Bio Protoc. 2023 Aug 5; 13(15): e4756.
Arend et al. (2023) Widening the landscape of transcriptional regulation of green algal photoprotection. Nat Commun. 2023 May 10;14(1):2687. doi: 10.1038/s41467-023-38183-4. PMID: 37164999
Makowski et al (2023) Activity modulation of the Escherichia coli F1FO ATP synthase by a designed antimicrobial peptide via cardiolipin sequestering
Cao et al. (2023). An unexpected hydratase synthesizes the green light--absorbing pigment fucoxanthin
Jin et al. (2023) Dual roles for CND1 in maintenance of nuclear and chloroplast genome stability in plants. Cell Rep. 2023 Mar 28;42(3):112268. doi: 10.1016/j.celrep.2023.112268. Epub 2023 Mar 17.
Ruiz-Sola et al. (2023) Light-independent regulation of algal photoprotection by CO2 availability. Nat Commun. 2023 Apr 8;14(1):1977. doi: 10.1038/s41467-023-37800-6.
von Bismarck, et al (2023). Light acclimation interacts with thylakoid ion transport to govern the dynamics of photosynthesis in Arabidopsis. New Phytol. 2023;237(1):160-176. doi:10.1111/nph.18538
Oliveira Souza et al. (2022) IMC10 and LMF1 mediate mitochondrial morphology through mitochondrion-pellicle contact sites in Toxoplasma gondii. J Cell Sci. 2022;135(22):jcs260083. doi:10.1242/jcs.260083
Baidukova et al. (2022) Gating and ion selectivity of Channelrhodopsins are critical for photo-activated orientation of Chlamydomonas as shown by in vivo point mutation. Nat Commun. 2022;13(1):7253. Published 2022 Nov 25. doi:10.1038/s41467-022-35018-6
Gao Y et al. (2022) Chloroplast translational regulation uncovers nonessential photosynthesis genes as key players in plant cold acclimation. Plant Cell. 2022 Apr 26;34(5):2056-2079. doi: 10.1093/plcell/koac056. PMID: 35171295; PMCID: PMC9048916.
Li et al. (2022) The CDC48 complex mediates ubiquitin-dependent degradation of intra-chloroplast proteins in plants. Cell Rep. 2022 Apr 12;39(2):110664. doi: 10.1016/j.celrep.2022.110664. PMID: 35417702.
Gruttner et al. (2022) The P-type pentatricopeptide repeat protein DWEORG1 is a non-previously reported rPPR protein of Arabidopsis mitochondria. Sci Rep. 2022 Jul 21;12(1):12492. doi: 10.1038/s41598-022-16812-0. PMID: 35864185; PMCID: PMC9304396.
Urban, Rogowski & Romanowska (2022), Crucial role of the PTOX and CET pathways in optimizing ATP synthesis in mesophyll chloroplasts of C3 and C4 plants, Environmental and Experimental Botany, Volume 202, October 2022, 105024,
Bru, Steen, Park, et al. (2022) The major trimeric antenna complexes serve as a site for qH-energy dissipation in plants. J Biol Chem. 2022;298(11):102519. doi:10.1016/j.jbc.2022.102519
Lim et al (2022). Arabidopsis guard cell chloroplasts import cytosolic ATP for starch turnover and stomatal opening. Nat Commun. 2022 Feb 3;13(1):652. doi: 10.1038/s41467-022-28263-2. PMID: 35115512; PMCID: PMC8814037.
Burlacot et al. (2022) Alternative photosynthesis pathways drive the algal CO2-concentrating mechanism. Nature 605, 366–371 (2022). https://doi.org/10.1038/s41586-022-04662-9
Bychkov et al. (2022) The role of PAP4/FSD3 and PAP9/FSD2 in heat stress responses of chloroplast genes. Plant Sci. 2022 Sep;322:111359. doi: 10.1016/j.plantsci.2022.111359. Epub 2022 Jun 20. PMID: 35738478.
Ripamonti et al (2022). Silencing of ATP Synthase Beta Impairs Egg Development in the Leafhopper Scaphoideus titanus, Vector of the Phytoplasma Associated with Grapevine Flavescence Doree. International Journal of Molecular Sciences. 2022; 23(2):765. https://doi.org/10.3390/ijms23020765
Tanno et al. (2021) The four-celled Volvocales green alga Tetrabaena socialis exhibits weak photobehavior and high-photoprotection ability. PLoS One. 2021 Oct 26;16(10):e0259138. doi: 10.1371/journal.pone.0259138. PMID: 34699573; PMCID: PMC8547699.
Mazur et al. (2021) The SnRK2.10 kinase mitigates the adverse effects of salinity by protecting photosynthetic machinery. Plant Physiol. 2021 Dec 4;187(4):2785-2802. doi: 10.1093/plphys/kiab438. PMID: 34632500; PMCID: PMC8644180.
Trojak et al. (2021) Effects of partial replacement of red by green light in the growth spectrum on photomorphogenesis and photosynthesis in tomato plants. Photosynth Res. 2021 Sep 27. doi: 10.1007/s11120-021-00879-3. Epub ahead of print. PMID: 34580802.
Choi et al. (2021) Augmented CO2 tolerance by expressing a single H+-pump enables microalgal valorization of industrial flue gas. Nat Commun. 2021 Oct 18;12(1):6049. doi: 10.1038/s41467-021-26325-5. PMID: 34663809; PMCID: PMC8523702.
von Bismarck et al. (2021) Light acclimation interacts with thylakoid ion transport to govern the dynamics of photosynthesis. Research Square; 2021. DOI: 10.21203/rs.3.rs-948381/v1.
Chen, Liu & Liu (2021) Loss-Function of EGY1 Results in Photosynthesis Damage through Reducing Stability of Photosystem II in Arabidopsis thaliana. Russ J Plant Physiol (2021).
Curien et al. (2021) Mixotrophic growth of the extremophile galdieria sulphuraria reveals the flexibility of its carbon assimilation metabolism. New Phytol. 2021 Mar 25. doi: 10.1111/nph.17359. Epub ahead of print. PMID: 33764540.
Maclean et al. (2021) Complexome profile of Toxoplasma gondii mitochondria identifies divergent subunits of respiratory chain complexes including new subunits of cytochrome bc1 complex. PLoS Pathog. 2021 Mar 2;17(3):e1009301. doi: 10.1371/journal.ppat.1009301. PMID: 33651838.
Picariello et al. (2020). TIM, a Targeted Insertional Mutagenesis Method Utilizing CRISPR/Cas9 in Chlamydomonas Reinhardtii. PLoS One. 2020 May 13;15(5):e0232594. doi: 10.1371/journal.pone.0232594.
Pattanaik et al. (2020). Introduction of a green algal squalene synthase enhances squalene accumulation in a strain of Synechocystis sp. PCC 6803. Metabolic Engineering Communications,Volume 10, June 2020, e00125
Mares et al. (2020). Hydrosoluble phylloplane components of Theobroma cacao modulate the metabolism of Moniliophthora perniciosa spores during germination.Fungal Biol. 2020 Jan;124(1):73-81. doi: 10.1016/j.funbio.2019.11.008.
Gabilly et al. (2019). Regulation of photoprotection gene expression in Chlamydomonas by a putative E3 ubiquitin ligase complex and a homolog of CONSTANS. Proc Natl Acad Sci U S A. 2019 Aug 12. pii: 201821689. doi: 10.1073/pnas.1821689116.
Vojta and Fulgosi (2019). Topology of TROL protein in thylakoid membranes of Arabidopsis thaliana. Physiol Plant. 2019 Jan 20. doi: 10.1111/ppl.12927.
Roth et al. (2019). Regulation of Oxygenic Photosynthesis during Trophic Transitions in the Green Alga Chromochloris zofingiensis. Plant Cell. 2019 Feb 20. pii: tpc.00742.2018. doi: 10.1105/tpc.18.00742.
Aihara et al. (2019). Algal photoprotection is regulated by the E3 ligase CUL4-DDB1DET1. Nat Plants. 2019 Jan;5(1):34-40. doi: 10.1038/s41477-018-0332-5.
Kong et al. (2018) Interorganelle Communication: Peroxisomal MALATE DEHYDROGENASE2 Connects Lipid Catabolism to Photosynthesis through Redox Coupling in Chlamydomonas. Plant Cell. 2018 Aug;30(8):1824-1847. doi: 10.1105/tpc.18.00361
Schottler et al. (2017). The plastid-encoded PsaI subunit stabilizes photosystem I during leaf senescence in tobacco. J Exp Bot. 2017 Feb 1;68(5):1137-1155. doi: 10.1093/jxb/erx009.
Jespersen et al. (2017). Metabolic Effects of Acibenzolar-S-Methyl for Improving Heat or Drought Stress in Creeping Bentgrass. Front Plant Sci. 2017 Jul 11;8:1224. doi: 10.3389/fpls.2017.01224. eCollection 2017. (western blot, Agostis stolonifera cv. ‘Penncross’)
Fristedt et al. (2015). The thylakoid membrane protein CGL160 supports CF1CF0 ATP synthase accumulation in Arabidopsis thaliana. PLoS One. 2015 Apr 2;10(4):e0121658. doi: 10.1371/journal.pone.0121658.
Rurek et al. (2015). Biogenesis of mitochondria in cauliflower (Brassica oleracea var. botrytis) curds subjected to temperature stress and recovery involves regulation of the complexome, respiratory chain activity, organellar translation and ultrastructure. Biochim Biophys Acta. 2015 Jan 21. pii: S0005-2728(15)00016-X. doi: 10.1016/j.bbabio.2015.01.005.
Eom et al. (2014). Bacillus subtilis HJ18-4 from Traditional Fermented Soybean Food Inhibits Bacillus cereus Growth and Toxin-Related Genes. J Food Sci. 2014 Nov;79(11):M2279-87. doi: 10.1111/1750-3841.12569. Epub 2014 Oct 30.
Lintala et al. (2013). Arabidopsis tic62 trol mutant lacking thylakoid bound ferredoxin-NADP+ oxidoreductase shows distinct metabolic phenotype. Mol Plant Sep 16.
Teng et al. (2013). Mitochondrial Genes of Dinoflagellates Are Transcribed by a Nuclear-Encoded Single-Subunit RNA Polymerase. PLOS ONE, June 2013. (immuofluorescence)
Rasala et al. (2013). Expanding the spectral palette of fluorescent proteins for the green microalga Chlamydomonas reinhardtii. Plant J. March 23.
Heinnickel et al. (2013). Novel thylakoid membrane greencut protein cpld38 impacts accumulation of the cytochrome b6f complex and associated regulatory processes. J. Biol. Chem. Jan 9. - Protocols
-
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extractsProtein Quantitation in plant and algal samples using Agrisera global antibodies
Methodology: Plant samples are generally ground with liquid nitrogen in a mortar and pestle. The resulting powder is transferred to a plastic tube. Algal samples can be either concentrated by centrifugation or, preferably, by filtration onto glass fiber filters. Solubilization is performed in Agrisera protein extraction buffer (PEB, AS08 300) containing 0.1mg/mL PefaBloc SC (AEBSF) protease inhibitor (Roche). Disruption is most optimally obtained through flash freezing of the sample in liquid nitrogen alternated with thawing by sonication with a microtip. This process can be repeated depending on the toughness of the sample. The sample is adjusted to 50 mM dithiothreitol and heated to 70°C for 5 minutes. Samples are cooled and centrifuged briefly prior to electrophoresis.
Optimal quantitation is achieved using moderate sample loads per gel lane, generally 0.5 to 2.5 ug total protein, depending on the abundance of the target protein.
Electrophoresis and Immunoblotting: Once solubilized, the proteins can be separated electrophoretically in a number of systems. We obtain optimal results with the Invitrogen NuPAGE gel system using Bis-Tris 4-12% gradient gels. Proteins are separated in MES SDS running buffer according to the manufacturer’s recommendations at 200 V for 35 minutes. The gels are transferred to PVDF in the same apparatus, the SureLock XCell blot module, for 60 minutes at 30 V for a single gel or 80 minutes for a pair. Following transfer the blots are blocked with non-fat dry milk up to 10 % in TBS-T, for 1 h/RT with gentle agitation. The blot is incubated with primary antibody, usually at 1:25 000 to 1:50 000, for 1 h/RT (if extreme femtogram detection reagents are used) or in lower primary antibody dilution for less sensitivie reagents (mid picogram and lower).
For quantitation a relatively high primary antibody: target protein ratio gives more reliable results than immunoblots at low ratios of primary antibody:target protein.
The blot is washed extensively in TBS-T (twice briefly, once for 15 minutes and three times for five minutes). The blot is incubated with secondary antibody, for example goat anti-rabbit IgG horse radish peroxidase conjugated, AS09 602 (Agrisera), for 1h/RT. The blot is washed as above and developed with ECL detection reagents.Quantitation: When quantitated standards are included on the blot, the samples can be quantitated using the available software. Excellent quantitation can be obtained with images captured on the Bio-Rad Fluor-S-Max or equivalent instrument using Bio-Rad QuantityOne software. The contour tool is used to select the area for quantitation and the values are background subtracted to give an adjusted volume in counts for each standard and sample. Using above protocol linear standard curves are generated over 1-1.5 orders of magnitude range in target load. It is important to note that immunodetections usually show a strongly sigmoidal signal to load response curve, with a region of trace detection of low loads, a pseudolinear range and a region of saturated response with high loads. For immunoquantitation it is critical that the target proteins in the samples and the standard curve fall within the pseudolinear range. Our total detection range using this protocol spans over 2 orders of magnitude, but the quantifiable range is narrower.
References:MacKenzie et al (2005). Large reallocations of carbon, nitrogen and photosynthetic reductant among phycobilisomes, photosystems and Rubisco during light acclimation in Synechococcus elongatus are constrained in cells under low environmental inorganic carbon. Arch of Microbiol. 183: 190 - 202.
Bouchard et al. (2006) UVB effects on the photosystem II-D1 protein of phytoplankton and natural phytoplankton communities. Photochem and Photobiol 82: 936-951.
Morash et al. (2007) Macromolecular dynamics of the photosynthetic system over a seasonal developmental progression in Spartina alterniflora. Can J. of Bot. 85: 476-483(8)Application example
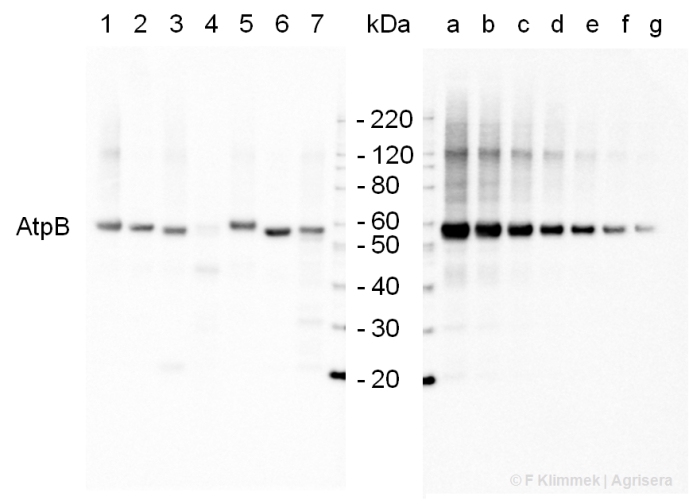
.jpg)
2 µg of total protein from (1-7) Prochlorococcus sp. extracted with Protein Extration Buffer, PEB (AS08 300), (8-11) Rubisco protein standard (AS01 017S) were separated on 4-12% NuPage (Invitrogen) LDS-PAGE and blotted 1h to PVDF. Blots were blocked immediately following transfer for 1h/RT with agitation. Blots were incubated in the primary antibody at a dilution of 1: 20 000 for 1h at room temperature with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, then washed once for 15 min and 3 times for 5 min in TBS-T at room temperature with agitation. Blots were incubated in secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated) diluted to 1:50 000 for 1h/RT with agitation. The blots were washed as above and developed for 5 min with ECL detection reagent according the manufacturers instructions. Images of the blots were obtained using a CCD imager (FluorSMax, Bio-Rad) and Quantity One software (Bio-Rad). There is a low level of a known RbcL cleavage product below the main band in some of the Prochlorococcus samples.
Recommended secondary antibodies: goat anti-rabbit HRP conjugated, goat anti-rabbit ALP conjugated
Recommended chemiluminescent detection reagent: AgriseraECLBright - Reviews:
-
Auroy Pascaline | 2022-02-24I use this antibody on Chlamydomonas, concentration 1/2500, on N deprivation condition , I see signal with 2.5µg of total protein.Manuela Leonardelli | 2019-06-18Loaded about 3 ug of protein from Arabidopsis thaliana seedlings. Signal is very clear and clean. No unspecific bands. Exposure time <1min.
Used as loading controlMichele Grieco | 2014-06-16For all these antibodies I loaded an amount of sample with a chlorophyll concentration of only 0,5 µl/ml, and in western blotting analyses the ECL signal was very intense. Moreover, I did not get aspecific bands for any of these antibodies.Andre Nordhues/Marita Hermann | 2012-11-2750 µg of total protein from Chlamydomonas reinhardtii was separated on 13 % SDS Gel and blotted 1h to PVDF.Blots were incubated in the primary antibody at a dilution of 1: 6 000 for 1h at room temperature with agitation.The primary antibody can be reused several times. Three bands are detected: 70 kDa (chloroplastic CF1 Atp Synthase), ~90 kDa, and ~120 kDa.
Accessories
AS03 030 | Clonality: Polyclonal | Host: Chicken | Reactivity: [global antibody] for plant and bacterial F-type ATP synthases
Benefits of using this antibody
AS05 085-10 | Clonality: Polyclonal | Host: Rabbit | Reactivity: [global antibody] for plant, green alga and bacterial F-type ATP synthases
Benefits of using this antibody
AS16 3976 | Clonality: Polyclonal | Host: Rabbit | Reactivity: Arabidopsis thaliana, Brassica oleracea var. botrytis cv. ’Diadom’, [compartment marker] of mitochondrial inner membrane
Benefits of using this antibody