1
Anti-AtpB | Beta subunit of ATP synthase (chloroplastic + mitochondrial) (chicken antibodies)
AS03 030 | Clonality: Polyclonal | Host: Chicken | Reactivity: [global antibody] for plant and bacterial F-type ATP synthases
Benefits of using this antibody
- Product Info
-
Immunogen: KLH-conjugated synthetic peptide derived from available plant, algal (chloroplastic and mitochondrial) and bacterial sequences of beta subunits of F-type ATP synthases, including Arabidopsis thaliana chloroplastic ATP synthase subunit beta UniProt: P19366, TAIR: AtCg00480 and Arabidopsis thaliana mitochondrial ATP synthase subunit beta-1, UniProt: P83483, TAIR: At5g08670 as well as Chlamydomonas reinhardtii, UniProt: P06541 and A8IQU3
Host: Chicken Clonality: Polyclonal Purity: Purified, total IgY (chicken egg yolk immunoglobulin) in PBS pH 8. Contains 0.02 % sodium azide. Format: Liquid Quantity: 100 µl Storage: Store at 2-8°C.; make aliquots to avoid working with a stock. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Immunolocalization (IL), Immunogold (IG), Western blot (WB) Recommended dilution: 1 : 500 for localization of native enzyme by immunogold (IL), 1 : 5 000-1 : 8 000 (WB) Expected | apparent MW: 53.9 kDa (Arabidopsis thaliana), 51.7 kDa (Synechocystis PCC 6803), 53.7 kDa (Spinacia oleracea)
- Reactivity
-
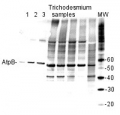
Confirmed reactivity: Arabidopsis thaliana, Hordeum vulgare, Phaeodactylum tricornutum, Spartina alterniflora, Spinacia oleracea, Synechocystis PCC 6803, Synechococcus PCC 7942, beef muscle, Rat liver Predicted reactivity: Acinetobacter baumannii, Bacillus subtilis, Brassica napus, Cannabis sativa, Clostridioides difficile, Cyanobacteria, E.coli K-12, Galdieria sulphuraria, Heliomicrobium modesticaldum Ice1, Manihot esculenta, Nicotiana plumbaginifolia, Saccharomyces cerevisiae, Salmonella typhimurium, Trichodesmium erythraeum, Triticum aestivum, Vitis vinifera, Zosteria marina, Yrsinia sp.
Species of your interest not listed? Contact usNot reactive in: Archeal V-type ATP synthase - Application Examples
-
application information 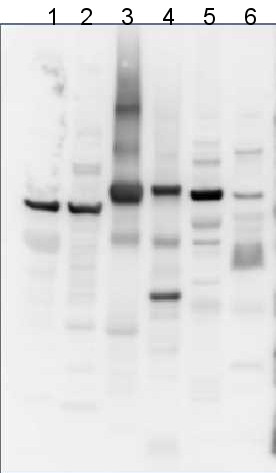
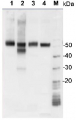
10 µg of total protein from samples such as beef muscle (1), rat liver (2), Arabidopsis thaliana leaf (3), Hordeum vulgare leaf (4), Synechocystis PCC 6803 total cell (5), Synechococcus PCC 7942 total cell (6) were extracted with Protein Extraction Buffer PEB (AS08 300). Samples were diluted with 1X sample buffer (NuPAGE LDS sample buffer (Invitrogen) supplemented with 50 mM DTT and heat at 70C for 5 min and keep on ice before loading. Protein samples were separated on Bolt 4-12% Tris-Bis gel (Invitrogen) LDS-PAGE and blotted for 1h to 1.5h on PVDF using Bolt Mini Blot Module. Blot was blocked immediately following transfer in 2% blocking reagent (GE RPN 2125; Healthcare) dissolved in 20 mM Tris, 137 mM sodium chloride pH 7.6 with 0.1% (v/v) Tween-20 (TBS-T) for 1h at room temperature with agitation. Blot was incubated in the primary antibody at a dilution of 1: 5 000 (in blocking reagent) for 1h at room temperature with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, and then washed 1x15 min and 3x5 min with TBS-T at room temperature with agitation. Blot was incubated in secondary antibody (anti-chicken IgY horse radish peroxidase conjugated) diluted to 1:20 000 in blocking reagent for 1h at room temperature with agitation. The blot was washed as above. Signals in the blot were detected using Lumigen ECL Ultra Reagent (Lumigen TMA-6, Lumigen), and visualized using the Molecular Imager VersaDoc MP 4000 System (Bio-Rad) and Quantity One software (Bio-Rad). Exposure time was 30 seconds.Application examples: 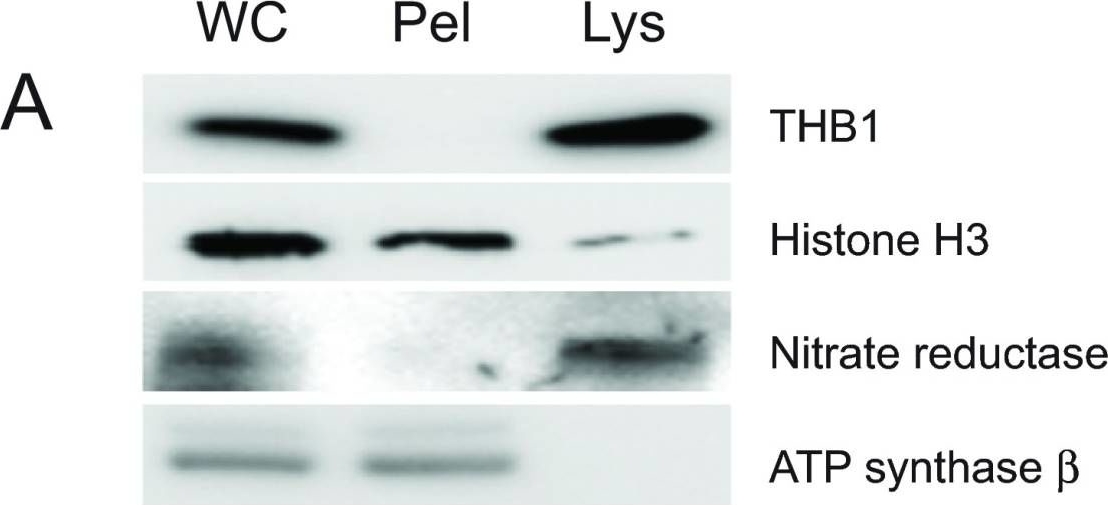
Reactant: Chlamydomonas reinhardtii (Green Alga)
Application: Western Blotting
Pudmed ID: 25653846
Journal: F1000Res
Figure Number: 1A
Published Date: 2015-02-06
First Author: Johnson, E. A. & Lecomte, J. T.
Impact Factor: None
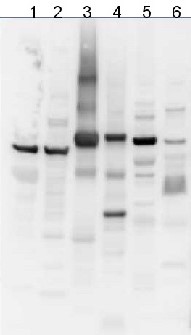
Open PublicationPurification of THB1 fromC. reinhardtii cell culture.Protein samples from different stages of purification were analyzed by electrophoresis. (A) Samples of protein extracts before and after lysis ofC. reinhardtii cells with liquid nitrogen. Proteins separated on 16.5% Tris-tricine gel then transferred to nitrocellulose and immunostained with antibodies against proteins localized to different cellular compartments. Lysis by liquid nitrogen enriches the lysate with soluble proteins without enrichment of proteins found in major algal organelles. Histone H3 protein is located in the nucleus, nitrate reductase is a soluble cytosolic protein and the ATP synthase ? subunit is part of the thylakoid membrane of the chloroplast. (B) Samples from different steps in the purification procedure were separated by electrophoresis. Following separation, the proteins within the gel were visualized using silver stain. (C) Proteins prepared identically to those detected in panel B were transferred to nitrocellulose followed by immunostaining with polyclonal antibodies specific for THB1. In addition to the protein samples, a lane was used for molecular weight markers (Spectra LR, ThermoScientific). The numbers indicated between the panels represents location of the markers (kDa molecular mass). WC, whole cell protein extract. Lys, protein extract lysate, and Pel, protein pellet following liquid nitrogen fracturing. QFF, concentrated sample following anion exchange chromatography. S75, concentrated sample following separation on the Superdex 75 column.
- Additional Information
-
Additional information: The anti-AtpB antibody will detect the mitochondrial form of the F1 ATP synthase subcomplex, as well as the chloroplastic CF1 ATP synthase and most known bacterial F-type ATP synthases, Peptide used for antibody production is located in a beta sheet, which is partly exposed near the surface of the AtpB protein Additional information (application): Results of immunogold studies using anti-AtpB antibody are published in Andersson et al. (2009).
- Background
-
Background: ATP synthase is the universal enzyme that synthesizes ATP from ADP and phosphate using the energy stored in a transmembrane ion gradient.
- Product Citations
-
Selected references: Neusius et al. (2022) Lysine acetylation regulates moonlighting activity of the E2 subunit of the chloroplast pyruvate dehydrogenase complex in Chlamydomonas. Plant J. 2022 Sep;111(6):1780-1800. doi: 10.1111/tpj.15924. Epub 2022 Aug 8. PMID: 35899410.
Levitan et al. (2019). Structural and functional analyses of photosystem II in the marine diatom Phaeodactylum tricornutum. Proc Natl Acad Sci U S A. 2019 Aug 27;116(35):17316-17322. doi: 10.1073/pnas.1906726116.
Nelson et al. (2019). Protein lysine methylation contributes to modulating the response of sensitive and tolerant Arabidopsis species to cadmium stress. doi: 10.1111/pce.13692.
Gellert et al. (2018). A single point mutation on the cucumber mosaic virus surface induces an unexpected and strong interaction with the F1 complex of the ATP synthase in Nicotiana clevelandii plants. Virus Res. 2018 Jun 2;251:47-55. doi: 10.1016/j.virusres.2018.05.005. (immunogold)
Quesada et al. (2011). Arabidopsis RUGOSA2 encodes an mTERF family member required for mitochondrion, chloroplast and leaf development. Plant J. Nov;68(4):738-53. doi: 10.1111/j.1365-313X.2011.04726.x. Epub 2011 Sep 13.
Andersson et. al (2009). Co-localization of P-glycerate kinase, P-ribulokinase, ADP-glucose pyrophosphorylase and Rubisco activase with CF1 in pea leaf chloroplasts. Plant Science 177:136-14. (immunologold). - Protocols
-
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extractsAtpB quantitation in plant and algal samples using Agrisera Anti-AtpB global antibody and AtpB protein standard
Methodology: Plant samples are generally ground with liquid nitrogen in a mortar and pestle. The resulting powder is transferred to a plastic tube. Algal samples can be either concentrated by centrifugation or, preferably, by filtration onto glass fiber filters. Solubilization is performed in Agrisera protein extraction buffer (PEB, AS08 300) containing 0.1mg/mL PefaBloc SC (AEBSF) protease inhibitor (Roche). Disruption is most optimally obtained through flash freezing of the sample in liquid nitrogen alternated with thawing by sonication with a microtip. This process can be repeated depending on the toughness of the sample. The sample is adjusted to 50 mM dithiothreitol and heated to 70°C for 5 minutes. Samples are cooled and centrifuged briefly prior to electrophoresis.Optimal quantitation is achieved using moderate sample loads per gel lane, generally 0.5 to 2.5 ug total protein, depending on the abundance of the target protein.
Electrophoresis and Immunoblotting: Once solubilized, the proteins can be separated electrophoretically in a number of systems. We obtain optimal results with the Invitrogen NuPAGE gel system using Bis-Tris 4-12% gradient gels. Proteins are separated in MES SDS running buffer according to the manufacturer’s recommendations at 200 V for 35 minutes. The gels are transferred to PVDF in the same apparatus, the SureLock XCell blot module, for 60 minutes at 30 V for a single gel or 80 minutes for a pair. Following transfer the blots are blocked with non-fat dry milk up to 10 % in TBS-T, for 1 h/RT with gentle agitation. The blot is incubated with primary antibody, usually at 1:25 000 to 1:50 000, for 1 h/RT (if extreme femtogram detection reagents are used) or in lower primary antibody dilution for less sensitivie reagents (mid picogram and lower).
For quantitation a relatively high primary antibody: target protein ratio gives more reliable results than immunoblots at low ratios of primary antibody:target protein.
The blot is washed extensively in TBS-T (twice briefly, once for 15 minutes and three times for five minutes). The blot is incubated with secondary antibody, for example goat anti-chicken IgY horse radish peroxidase conjugated, for 1h/RT. The blot is washed as above and developed with ECL detection reagents.Quantitation: When quantitated standards are included on the blot, the samples can be quantitated using the available software. Excellent quantitation can be obtained with images captured on the Bio-Rad Fluor-S-Max or equivalent instrument using Bio-Rad QuantityOne software. The contour tool is used to select the area for quantitation and the values are background subtracted to give an adjusted volume in counts for each standard and sample. Using above protocol linear standard curves are generated over 1-1.5 orders of magnitude range in target load. It is important to note that immunodetections usually show a strongly sigmoidal signal to load response curve, with a region of trace detection of low loads, a pseudolinear range and a region of saturated response with high loads. For immunoquantitation it is critical that the target proteins in the samples and the standard curve fall within the pseudolinear range. Our total detection range using this protocol spans over 2 orders of magnitude, but the quantifiable range is narrower.
Recommended secondary antibody from Agrisera: Rabbit anti-chicken HRP conjugated or Goat anti-chicken HRP conjugated or Goat anti-chicken ALP conjugated
Recommended chemiluminescent detection reagent: AgriseraECLBrightWestern Blot using IgY
Important note: Please, be aware, that often protocols used for IgG antibodies (rabbit, goat, mice) have to be optimised to get best results with IgY antibodies.1. Block unspecific binding by washing membrane in 25 ml blocking buffer/13x13 cm filer for 2 hours at room temperature on a shaker.
* Example of a Blocking buffer (Blotto)
1x TBS (20 mM Tris/HCl, pH 7.5,150 mM NaCl, 3-5 % low-fat dried milk powder, 0.05 % Tween-20 (or Nonidet P-40)
Blotto - Limitations:
o Usually very effective however: there are complex carbohydrates in milk and these will absorb out antibodies that recognize carbohydrate determinants.
o Some IgY antibodies might recognize milk proteins (high background signal).
o 10 % nonfat dry milk might block so efficiently, and in some cases so well, that no bands of interest will be seen.
Blocking insufficient - alternatives to try:Increase non-fat milk to 10-15 % 1h/RT incubation.
2. Wash each filter briefly, twice, then 1 x 15 minutes and 4 x 5 minutes at RT in Washing buffer. In case of still high background siganl - increase the length of a washing step even more than what is recommended above.
o Washing buffer:
1 x TBS, 0.05 % Tween-203. Add primary antibody to 10-20 ml of Antibody buffer (dilution range from 1:100 to 1: 30 000) and incubate the filter for 1-3 hours at room temperature (or 37°C, optional).
o Antibody buffer: 1 x TBS, 2% milk powder, 0.05 % Tween 204. Rinse the filter briefly twice, following by 1 x 15 and 4 x 5 minutes at RT in Washing buffer.
5. Add secondary antibody in Antibody Buffer. Use for instance rabbit or goat anti-IgY HRP conjugated. Start with the dilution at least 1 :10 000 (even higher dilutions are recommended).
Cross-reactivity signal coming from a secondary antibody can be easily checked by omitting a primary antibody in the whole procedure. Recommended secondary antibody from Agrisera: Rabbit anti-chicken HRP conjugated or Goat anti-chicken HRP conjugated or Goat anti-chicken ALP conjugated6. Wash the filter 4 x 5 minutes in Washing buffer, followed by 1 x 15 minutes in dH2O at room temperature.
Important!
Amount of membrane washes. In case of high unspecific background, increase the time of the wash with frequent exchange of Washing buffer.
In case of high background levels, firstly test if the secondary antibody is not contributing to the background. Use a small piece of empty transfer membrane, follow the procedure above without the step with primary antibodies. Recommended chemiluminescence detection reagent in mid picogram range: AgriseraECLBright, since primary antibodies can be used in a very high dilution, often background signal will be diluted out at that step.
Using extra sensitivite development systems (above extreme femtogram) can contribute to increased background signals.
Check also: Western Blot troubleshootingor contact Agrisera Technical Support or talk with us directly on the website (bottom right corner).
ImmunoGold
Gellért et al. (2018). A single point mutation on the cucumber mosaic virus surface induces an unexpected and strong interaction with the F1 complex of the ATP synthase in Nicotiana clevelandii plants. Virus Res. 2018 Jun 2;251:47-55. doi: 10.1016/j.virusres.2018.05.005.Andersson et. al (2009). Co-localization of P-glycerate kinase, P-ribulokinase, ADP-glucose pyrophosphorylase and Rubisco activase with CF1 in pea leaf chloroplasts. Plant Science 177:136-14. (immunologold)
- Reviews:
-
Yu Qing-Bo | 2009-05-07This antibody works very well in our lab, when it was used to detect the accumulation of photosynthetic protein in Arabidopsis