1
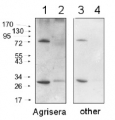
Anti-DHAR1 | Dehydroascorbate Reductase 1
AS11 1746 | Clonality: Polyclonal | Host: Rabbit | Reactivity: Arabidopsis thaliana, Solanum lycopersicum
- Product Info
-
Immunogen: KLH-conjugated synthetic peptide derived from known DHAR1 sequence of Arabidopsis thaliana Q9FWR4, At1g19570
Host: Rabbit Clonality: Polyclonal Purity: Immunogen affinity purified serum in PBS pH 7.4. Format: Lyophilized Quantity: 200 µg Reconstitution: For reconstitution add 200 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Western blot (WB) Recommended dilution: 1 : 5000 (WB) Expected | apparent MW: 23.6 | 23.4 kDa - Reactivity
-
Confirmed reactivity: Arabidopsis thaliana, Kandelia candel, Solanum lycopersicum
Predicted reactivity: Nicotiana tabacum, Populus trichocarpa, Ricinus communis, Solanum tuberosum, Triticum aestivum, Zinnia elegans
Species of your interest not listed? Contact usNot reactive in: No confirmed exceptions from predicted reactivity are currently known - Application Examples
-
application example
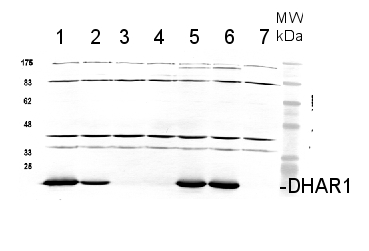
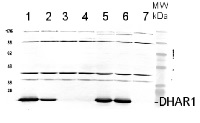
1cm2 of a leaf from Arabidopsis thaliana Col-0 (1) and or t-DNA insertion lines dhar1-1 (2), dhar1-2 (3), dhar1-3 (4), dhar2-1 (5), dhar2-2 (6), dhar1-3 EOS-DHAR1 (7), was extracted using 200µl Lyse&Load-Buffer (Grefen et al. 2009). 10 µl were separated on a 15% SDS-PAGE and blotted 1h to PVDF (using Bjerrum Buffer in a semidry blot). Blots were blocked with 5% Milk in 1xTBS-Tween20 (1%) for 1h at room temperature (RT) with agitation. Blot was incubated in the primary antibody at a dilution of 1:5000 (in 5% Milk 1xTBS-Tween20 (1%) + 0.01 % NaN3) ON at 4°C with agitation. The antibody solution was decanted and the blot was washed 3 times for 10 minutes with 1x TBS-Tween20 at RT with agitation. Blot was incubated in secondary antibody BioRad anti-rabbit IgG AP-conjugate (#170-6518) diluted to 1:2000 in 5% Milk 1xTBS-Tween20 (1%) + 0.01 % NaN3 for 1h at RT with agitation. The blot was washed as above, equilibrated in staining buffer (100mM Tris-HCl, 100mM NaCl, 5mM MgCl2, see Grefen et al. 2009) and developed for 5-15 min. with staining solution (Nitro blue tetrazolium chloride (NBT) and 5-bromo-4-chloro-3-indoylphosphate-p-toluidin (BCIP) in staining buffer).
Courtesy Dr. Chrisopher Grefen, UK
Application examples: 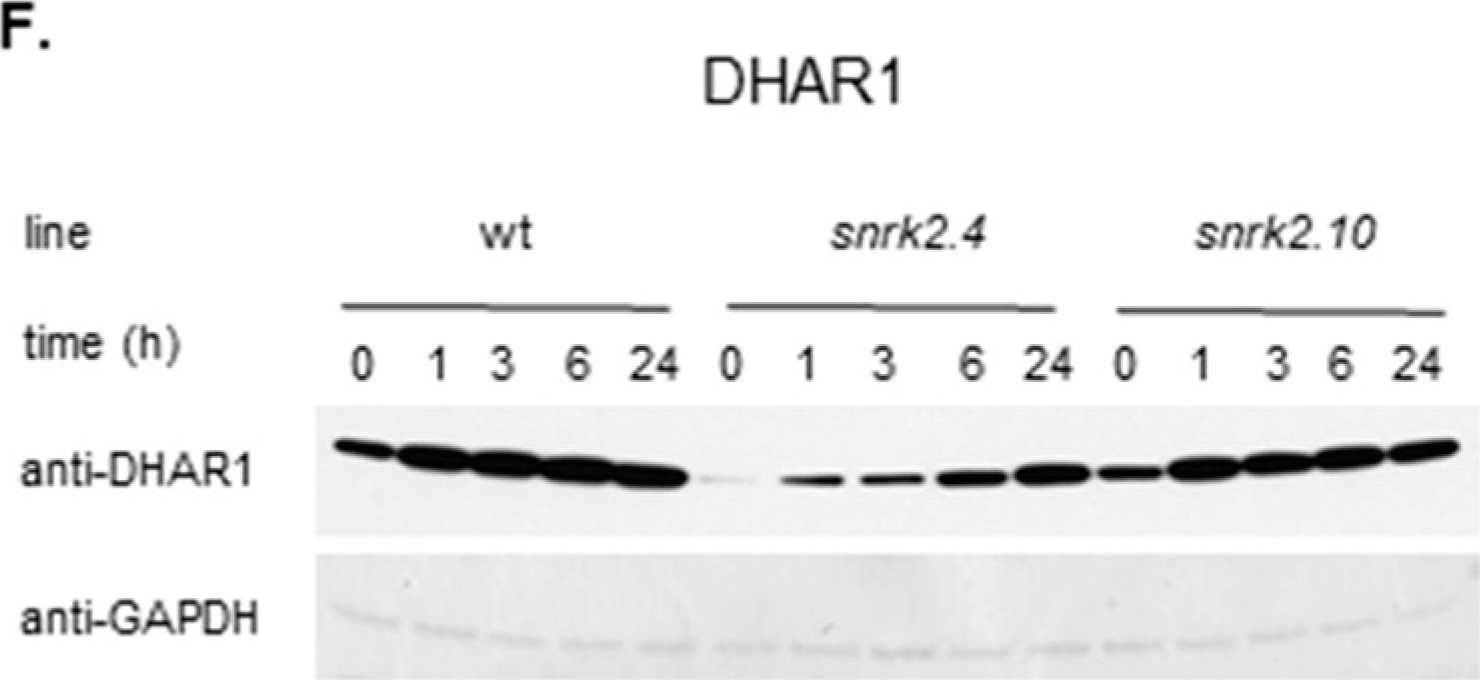
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 30609769
Journal: Int J Mol Sci
Figure Number: 4F
Published Date: 2019-01-02
First Author: Szyma?ska, K. P., Polkowska-Kowalczyk, L., et al.
Impact Factor: 5.542
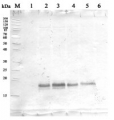
Open PublicationSnRK2.4 and SnRK2.10 regulate enzymes of the ascorbate cycle during response to salt stress. Wild type and snrk2 mutant seedlings were subjected to treatment with 150 mM NaCl for times indicated. Expression of (A) APX1—Ascorbate Peroxidase 1, (B) APX2—Ascorbate Peroxidase 2, (C) APX6—Ascorbate Peroxidase 6, and (D) DHAR1—Dehydroascorbate Reductase 1 was monitored by RT-qPCR; error bars represent SD; stars represent statistically significant differences in comparison with the wt plants (Student t-test) where * p < 0.05; ** p < 0.001; *** p < 0.0001. Total protein level of (E) APX and (F) DHAR1 was monitored with immunoblot analysis; after exposure, membranes were stripped and reused for GAPDH detection as a loading control. At least two independent biological replicates of the experiment were performed. Results of one representative experiment are shown.
- Background
-
Background: DHAR1 ( Dehydroascorbate Reductase 1) the protein is induced by jasmonic acid and oxidative chemical stresses and is a key component of the ascorbate recycling system. Involved in redox homeostasis under biotic and abiotic inducers. Localized in mitochondria. Synonymes: glutathione-dependent dehydroascorbate reductase 1, chloride intracellular channel homolog 1, CLIC homolog 1, glutathione-dependent dehydroascorbate reductase 1, AtDHAR1, GSH-dependent dehydroascorbate reductase 1, mtDHAR, AT1G19570.
- Product Citations
-
Selected references: Szymańska et al. (2019). SNF1-Related Protein Kinases SnRK2.4 and SnRK2.10 Modulate ROS Homeostasis in Plant Response to Salt Stress. Int J Mol Sci. 2019 Jan 2;20(1). pii: E143. doi: 10.3390/ijms20010143.
Witzel et al. (2017). Temporal impact of the vascular wilt pathogen Verticillium dahliae on tomato root proteome. J Proteomics. 2017 Oct 3;169:215-224. doi: 10.1016/j.jprot.2017.04.008.
Wang et al. (2014). Proteomic analysis of salt-responsive proteins in the leaves of mangrove Kandelia candel during short-term stress. PLoS One. 2014 Jan 8;9(1):e83141. doi: 10.1371/journal.pone.0083141. eCollection 2014. - Protocols
-
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extracts
Oxygenic photosynthesis poster by prof. Govindjee and Dr. Shevela
Z-scheme of photosynthetic electron transport by prof. Govindjee and Dr. Björn and Dr. Shevela - Reviews:
-
This product doesn't have any reviews.