1
Anti-RbcL | Rubisco large subunit, form I (chicken)
AS01 017 | Clonality: Polyclonal | Host: Hen | Reactivity: [global antibody] for higher plants, algae, cyanobacteria | cellular [compartment marker] of plastid stroma
Benefits of using this antibody
- Product Info
-
Immunogen: KLH-conjugated synthetic peptide derived from all known plant,algal and cyanobacterial RbcL (Rubisco large subunit of Rubisco Form I) sequences, including Arabidopsis thaliana UniProt: O03042, TAIR: AtCg00490, Synechococcus sp. Q3ALL1
Host: Chicken Clonality: Polyclonal Purity: Purified, total IgY (chicken egg yolk immunoglobulin) in PBS pH 8. Contains 0.02 % sodium azide. Format: Liquid Quantity: 50 µl Storage: Store at 4°C; make aliquots to avoid working with a stock. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Immunogluorescence (IF), Immunolocalization (IL), ImmunoGold (IG), Western blot (WB) Recommended dilution: (IL) tested on a grass species, formaldehyde-fixed and paraffin-embedded tissue following the protocol from Gonzalez et al, (1998) Plant Physiol, V, 116, 1 : 10 000-1 : 20 000, 2 µg of total cellular protein, (WB) Expected | apparent MW: 52.7 kDa (Arabidopsis thaliana), 52.5 kDa (cyanobacteria), 52.3 kDa (Chlamydomonas reinhardtii)
- Reactivity
-
Confirmed reactivity: Arabidopsis thaliana, Fragilariopsis cylindrus, Lobaria pulmonaria, Medicago sativa, mixed phytoplankton, Microcystis aeruginosa PCC7806, Pinus strobus, Pisum sativum, Solanum tuberosum, Spartina alterniflora, Spinacia oleracea, Synechococcus sp. PCC7842, Thiobacillus sp. Ulmus sp. Predicted reactivity: Algae, Dicots, Conifers, Liverworts, Mosses, Prochlorophytes, Welwitschia Not reactive in: No confirmed exceptions from predicted reactivity are currently known - Application Examples
-
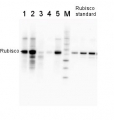
Application example 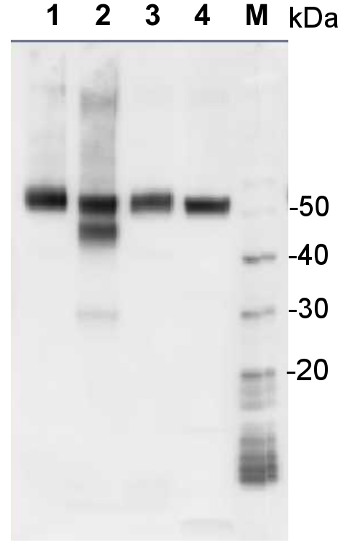
1 µg of total protein from samples such as Arabidopsis thaliana leaf (1) , Hordeum vulgare leaf (2), Zea mays leaf (3), Chlamydomonas reinhardtii total cell (4), were extracted with Protein Extraction Buffer PEB (AS08 300). Samples were diluted with 1X sample buffer (NuPAGE LDS sample buffer (Invitrogen) supplemented with 50 mM DTT and heat at 70°C for 5 min and keept on ice before loading. Protein samples were separated on 4-12% Bolt Plus gels, LDS-PAGE and blotted for 70 minutes to PVDF using tank transfer. Blots were blocked immediately following transfer in 2% blocking reagent (GE RPN 2125; Healthcare) or 5% non-fat milk dissolved in 20 mM Tris, 137 mM sodium chloride pH 7.6 with 0.1% (v/v) Tween-20 (TBS-T) for 1h at room temperature with agitation. Blots were incubated in the primary antibody at a dilution of 1: 10 000 (in blocking reagent) for 1h at room temperature with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, and then washed 1x15 min and 3x5 min with TBS-T at room temperature with agitation. Blots were incubated in secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated, secondary antibody AS10 1489, Agrisera) diluted to 1:25 000 in blocking reagent for 1h at room temperature with agitation. The blots were washed as above. The blot was developed for 5 min with chemiluminescent detection reagent according the manufacturers instructions. Images of the blots were obtained using a CCD imager (VersaDoc MP 4000) and Quantity One software (Bio-Rad). Exposure time was 30 seconds.Application examples: 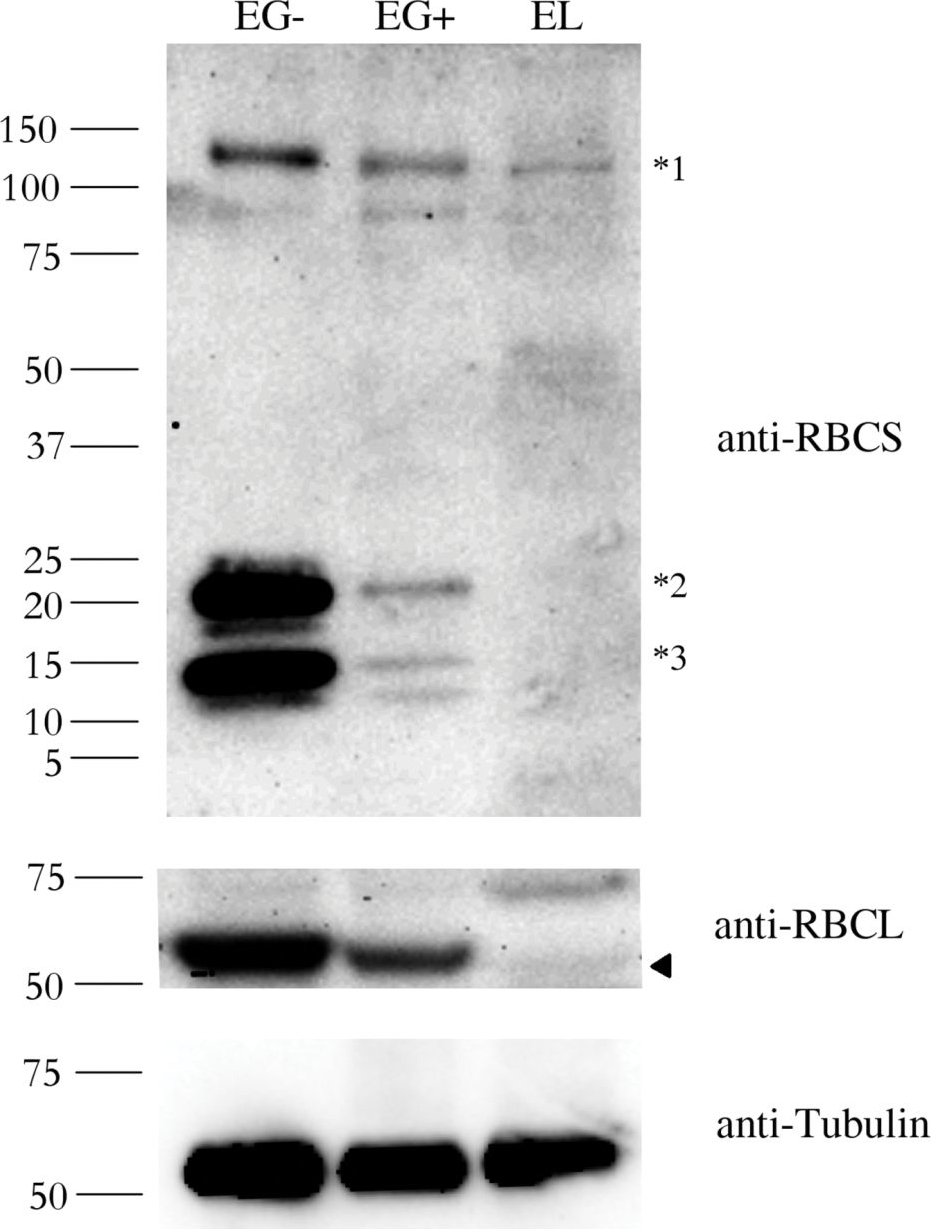
Reactant: Euglena
Application: Western Blotting
Pudmed ID: 27391690
Journal: PLoS One
Figure Number: 2A
Published Date: 2016-07-09
First Author: Záhonová, K., Füssy, Z., et al.
Impact Factor: 2.942
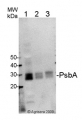
Open PublicationAbundance of the RBCS and RBCL proteins in Euglena gracilis and Euglena longa.Protein immunodetection was performed using anti-RBCS, anti-RBCL, and anti-Tubulin antibodies. Three bands with different molecular weights were observed in anti-RBCS immunoblotting. The ~130 kDa band (marked *1) corresponds to polyprotein synthesized in the nucleus. The ~15 kDa band (marked *3) corresponds to the processed monomer after cleavage of the signal sequence and excision of decapeptides. The ~22 kDa band (marked *2) possibly corresponds to a monomer still attached to the transit peptide. The identity of the RBCL protein (arrowhead in the anti-RBCL panel) was confirmed by mass-spectrometry. Tubulin served as a loading control. Molecular weights in kDa are indicated on the left. EG-, E. gracilis cultivated photosynthetically (without ethanol); EG+, E. gracilis cultivated mixotrophically (with ethanol); EL, E. longa.
Reactant: Euglena
Application: Western Blotting
Pudmed ID: 27391690
Journal: PLoS One
Figure Number: 4A,B,C
Published Date: 2016-07-09
First Author: Záhonová, K., Füssy, Z., et al.
Impact Factor: 2.942
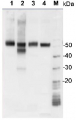
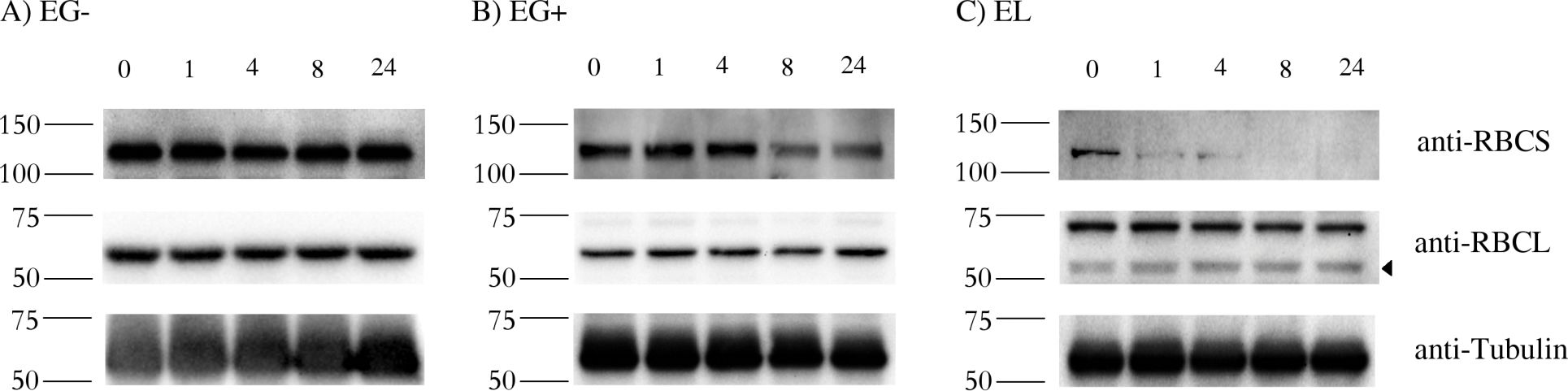
Open PublicationStability of RBCS and RBCL proteins in Euglena gracilis and Euglena longa.Cell cultures were treated with 20 ?g/ml of cycloheximide, aliquots were taken at 0, 1, 4, 8, and 24 h post treatment, and analyzed by western blotting using anti-RBCS, anti-RBCL and anti-Tubulin antibodies. Molecular weights (in kDa) are indicated on the left of each panel. The identity of the RBCL protein (arrowhead in the anti-RBCL panel) was confirmed by mass-spectrometry. Tubulin served as a loading control. Cultivation conditions and species are denoted as in Fig 2.
- Additional Information
-
Additional information: Peptide target used to elicit this antibody is not conserved in type II Rubisco found in dinoflagellates and some photosynthetic bacteria Additional information (application): This antibody detects RbcL protein from 102.6 fmoles and has been used as a control to ensure adequate permeabilization and fixation of toxic cyanobacterial cells in immunolabeling experiments (method based on: Orellana & Perry (1995) J Phycol 31: 785-794).
Antibody has been used in immunolabelling of intact cyanobacterial cells fixed with ethanol using a secondary anti-IgY antibody conjugated with a fluorochrome.
For Rubisco quantification using quantitative western blot technique, anti-RbcL antibody, (AS03 037) combined with Rubisco ready to use standard (AS01 017) is recommended. - Background
-
Background: Rubisco (Ribulose-1,5-bisphosphate carboxylase/oxygenase) catalyzes the rate-limiting step of CO2 fixation in photosynthesis. It is one of the most abundant proteins on Earth and its homology has been demonstrated from purple bacteria to flowering plants.
- Product Citations
-
Selected references: Guljamow et al. (2021) Diel Variations of Extracellular Microcystin Influence the Subcellular Dynamics of RubisCO in Microcystis aeruginosa PCC 7806. Microorganisms. 2021 Jun 10;9(6):1265. doi: 10.3390/microorganisms9061265. PMID: 34200971; PMCID: PMC8230624. (IF)
Morin et al. (2019). Morin et al. (2019). Response of the sea-ice diatom Fragilariopsis cylindrus to simulated polar night darkness and return to light. Limnology and Oceanography. 9999, 2019, 1–20. (sea-ice diatom)
Lv et al. (2019). Uncoupled Expression of Nuclear and Plastid Photosynthesis-Associated Genes Contributes to Cell Death in a Lesion Mimic Mutant. Plant Cell. 2019 Jan;31(1):210-230. doi: 10.1105/tpc.18.00813.
Gellért et al. (2018). A single point mutation on the cucumber mosaic virus surface induces an unexpected and strong interaction with the F1 complex of the ATP synthase in Nicotiana clevelandii plants. Virus Res. 2018 Jun 2;251:47-55. doi: 10.1016/j.virusres.2018.05.005.
Robert et al. (2015). Leaf proteome rebalancing in Nicotiana benthamiana for upstream enrichment of a transiently expressed recombinant protein. Plant Biotechnol J. 2015 Aug 19. doi: 10.1111/pbi.12452.
Morash et al. (2007). Macromolecular dynamics of the photosynthetic system over a seasonal developmental progression in Spartina alterniflora. Canadian J. of Botany, 2007, 85(5): 476-483, 10.1139/B07-043.
MacKenzie et al. (2005). Inorganic carbon acclimation in Synechococcus elongatus alters the dynamics of macromolecular pooks and photosynthetic fluxes in response to increased light. Photosynt Research 85: 341-357.
Schofield et al. (2003). Changes in macromolecular allocation in nondividins algal symbionts allow for photosynthetic acclimation in the lichen Lobaria pulmonaria. New Phytol 159: 709-718. - Protocols
-
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extractsRubisco quantitation in plant and algal samples using Agrisera anti-RbcL global antibody and Rubisco protein standard
Methodology: Plant samples are generally ground with liquid nitrogen in a mortar and pestle. The resulting powder is transferred to a plastic tube. Algal samples can be either concentrated by centrifugation or, preferably, by filtration onto glass fiber filters. Solubilization is performed in Agrisera protein extraction buffer (PEB, AS08 300) containing 0.1mg/mL PefaBloc SC (AEBSF) protease inhibitor (Roche). Disruption is most optimally obtained through flash freezing of the sample in liquid nitrogen alternated with thawing by sonication with a microtip. This process can be repeated depending on the toughness of the sample. The sample is adjusted to 50 mM dithiothreitol and heated to 70°C for 5 minutes. Samples are cooled and centrifuged briefly prior to electrophoresis.
Optimal quantitation is achieved using moderate sample loads per gel lane, generally 0.5 to 2.5 ug total protein, depending on the abundance of the target protein.
Electrophoresis and Immunoblotting: Once solubilized, the proteins can be separated electrophoretically in a number of systems. We obtain optimal results with the Invitrogen NuPAGE gel system using Bis-Tris 4-12% gradient gels. Proteins are separated in MES SDS running buffer according to the manufacturer’s recommendations at 200 V for 35 minutes. The gels are transferred to PVDF in the same apparatus, the SureLock XCell blot module, for 60 minutes at 30 V for a single gel or 80 minutes for a pair. Following transfer the blots are blocked with non-fat dry milk up to 10 % in TBS-T, for 1 h/RT with gentle agitation. The blot is incubated with primary antibody, usually at 1:25 000 to 1:50 000, for 1 h/RT (if extreme femtogram detection reagents are used) or in lower primary antibody dilution for less sensitivie reagents (mid picogram and lower).
For quantitation a relatively high primary antibody: target protein ratio gives more reliable results than immunoblots at low ratios of primary antibody:target protein.
The blot is washed extensively in TBS-T (twice briefly, once for 15 minutes and three times for five minutes). The blot is incubated with secondary antibody, for example goat anti-rabbit IgG horse radish peroxidase conjugated, AS09 602 (Agrisera), for 1h/RT. The blot is washed as above and developed with ECL detection reagents.Quantitation: When quantitated standards are included on the blot, the samples can be quantitated using the available software. Excellent quantitation can be obtained with images captured on the Bio-Rad Fluor-S-Max or equivalent instrument using Bio-Rad QuantityOne software. The contour tool is used to select the area for quantitation and the values are background subtracted to give an adjusted volume in counts for each standard and sample. Using above protocol linear standard curves are generated over 1-1.5 orders of magnitude range in target load. It is important to note that immunodetections usually show a strongly sigmoidal signal to load response curve, with a region of trace detection of low loads, a pseudolinear range and a region of saturated response with high loads. For immunoquantitation it is critical that the target proteins in the samples and the standard curve fall within the pseudolinear range. Our total detection range using this protocol spans over 2 orders of magnitude, but the quantifiable range is narrower.
References:
MacKenzie et al (2005). Large reallocations of carbon, nitrogen and photosynthetic reductant among phycobilisomes, photosystems and Rubisco during light acclimation in Synechococcus elongatus are constrained in cells under low environmental inorganic carbon. Arch of Microbiol. 183: 190 - 202.
Bouchard et al. (2006) UVB effects on the photosystem II-D1 protein of phytoplankton and natural phytoplankton communities. Photochem and Photobiol 82: 936-951.
Morash et al. (2007) Macromolecular dynamics of the photosynthetic system over a seasonal developmental progression in Spartina alterniflora. Can J. of Bot. 85: 476-483(8)Application example
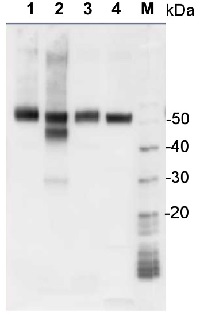
.jpg)
2 µg of total protein from (1-7) Prochlorococcus sp. extracted with Protein Extration Buffer, PEB (AS08 300), (8-11) Rubisco protein standard (AS01 017S) were separated on 4-12% NuPage (Invitrogen) LDS-PAGE and blotted 1h to PVDF. Blots were blocked immediately following transfer for 1h/RT with agitation. Blots were incubated in the primary antibody at a dilution of 1: 20 000 for 1h at room temperature with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, then washed once for 15 min and 3 times for 5 min in TBS-T at room temperature with agitation. Blots were incubated in secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated) diluted to 1:50 000 for 1h/RT with agitation. The blots were washed as above and developed for 5 min with ECL detection reagent according the manufacturers instructions. Images of the blots were obtained using a CCD imager (FluorSMax, Bio-Rad) and Quantity One software (Bio-Rad). There is a low level of a known RbcL cleavage product below the main band in some of the Prochlorococcus samples.
Recommended secondary antibodies: goat anti-rabbit HRP conjugated, goat anti-rabbit ALP conjugated
Recommended chemiluminescent detection reagent: AgriseraECLBrightMethod for embedding in LR WHITE for immunolabelling using anti-RbcL antibody
Prins et al. (2008). Cysteine proteinases regulate chloroplast protein content and composition in tobacco leaves:a model for dynamic interactions with ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) vesicular bodies. J.Ex.Bot.59:1935-1950. Method described below is a courtesy of Dr Aranda
Samples must be sectioned at 1 x 1 mm. 1)Fixation: Glutaraldehyde 0.5% + paraformaldehyde 2.5% in sodium phosphate buffer 0.1M pH= 7.2, 1 hour and 30 min. 2) 3 washes in sodium phosphate buffer 0.1M pH= 7.2 (15 min. each one). 3)Dehydratation and embedding:
-35% Ethanol……30 min.
-50% Ethanol……30 min.
-70% Ethanol……30 min.
-96% Ethanol……30 min.
-100% Ethanol……30 min.
-100% Ethanol……30 min.
-100% Ethanol……30 min.-75% Ethanol + 25% LR White…… 1 hour
-50% Ethanol + 50% LR White…… 1 hour
-25% Ethanol + 75% LR White…… 1 hour
-100% LR White…………………... 1 hour
-100% LR White…………………... overnight.4) Polymerization: Oven 12-18 hours at 50ºC. For flat molds in continous flux of N2
5) Block sectioning: Ultrathin sections (60-70nm) using a ultramicrotome. Ultrathin sections should be placed on formvar coated níkel grids.
IMMUNOLABELLING
1) Blocking: Place grids with sections into 30-μL drop of blocking buffer during 30 minutes. (Blocking buffer: phosphate-buffered saline (PBS) + 5% bovine serum albumin (BSA).
2) Incubation: Incubate in drop of primary antibody, anti-RbcL (Rubisco form I and II, Agrisera) diluted 1:250 in blocking buffer. 3 hours at room temperature (could be extend to overninght).
3) Rinse: Pass the grids through a series of drops of PBS + 1%BSA. Two or three changes during 5 minutes each one.
4) Incubation: Incubate grids in 30-μL of secondary antibody gold-labelled (British BioCell International, 10nm), diluted 1:50 in blocking buffer during 1.5 hours at room temperature.
5) Rinse: Pass the grids through a series of drops of PBS + 1%BSA. Two changes during 5 minutes each one.
6) Rinse: Pass the grids through a series of drops of PBS. Two or three changes during 5 minutes each one.
7) Rinse: Pass the grids through a series of drops of water (ultra-pure water must filtered 0.2 μm) . Four to five changes during 5 minutes each one, allowed to dry for later observation.
8) Staining: The grids may be post-stained with uranyl acetate and lead citrate.
Pao et al. (2018). Lamelloplasts and minichloroplasts in Begoniaceae: iridescence and photosynthetic functioning. J Plant Res. 2018 Mar 2. doi: 10.1007/s10265-018-1020-2. (ImmunoGold)
Liang et al. (2014). Cyanophycin mediates the accumulation and storage of fixed carbon in non-heterocystous filamentous cyanobacteria from coniform mats. PLoS One. 2014 Feb 7;9(2):e88142. doi: 10.1371/journal.pone.0088142. eCollection 2014. (ImmunoGold) - Reviews:
-
This product doesn't have any reviews.
Accessories
AS03 037 | Clonality: Polyclonal | Host: Rabbit | Reactivity: global antibody and compartment marker for higher plants, lichens, algae, cyanobacteria, dinoflagellates, diatoms
Benefits of using this antibody