3

Anti-RbcL | Rubisco large subunit, form I (rabbit)
AS03 037 | Clonality: Polyclonal | Host: Rabbit | Reactivity: global antibody and compartment marker for higher plants, lichens, algae, cyanobacteria, dinoflagellates, diatoms
Benefits of using this antibody
- Product Info
-
Immunogen: KLH-conjugated synthetic peptide conserved across all known plant, algal and cyanobacterial RbcL protein sequences (form I L8S8 and form II L2), including, Arabidopsis thaliana O03042, Hordeum vulgare P05698, Oryza sativa P0C510, Chlamydomonas reinhardtii P00877, Synechococcus PCC 7920 A5CKC5
Host: Rabbit Clonality: Polyclonal Purity: Serum Format: Lyophilized Quantity: 50 µl Reconstitution: For reconstitution add 50 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Immunofluorescence/confocal Immunolocalization (IL) (IF), Immunogold (IG), Tissue Printing (TP), Western blot (WB) Recommended dilution: Immunofluorescence/confocal microscopy (IF), 1: 1000 (IG), 1: 250 for images see Prins et al. (2008), detailed protocol available on request, 1: 800 (TP), 1: 5000 - 10 000 and more depending upon protein load and detection method (WB) Expected | apparent MW: 52.7 kDa (Arabidopsis thaliana), 52.5 kDa (cyanobacteria), 52.3 (Chlamydomonas reinhardtii)
- Reactivity
-
Confirmed reactivity: Agostis stolonifera cv. Penncross, Arabidopsis thaliana, Apium graveolens, Artemisia annua, Atrichum undulatum, Attheya longicornis, Baculogypsina sphaerulata (benthic foraminifer), Beta vulgaris, Begonia sp., Bienertia sinuspersici, Brassica napus, Kandelia candel, Cannabis sativa L., Chaetoceros furcellatus, Chlorococcum dorsiventrale, Colobanthus quitensis, Cicer arietinum, Chenopodium quinoa, Chlamydomonas raudensis, Chlamydomonas reinhardtii, Colobanthus quitensis Kunt Bartl, Chlorella sorokiniana, Chlorella vulgaris, Coscinodiscus concinnus, Cyanophora paradoxa, Cylindrospermopsis raciborskii CS-505, Cynara cardunculus, Emiliana huxleyi, Euglena gracilis, Ficus carica, Fortunella margarita Swingle, Fraxinus mandshurica, Fucus vesiculosus, Gladieria sulphuraria, Glycine max, Gonyaulax polyedra, Gongolaria barbata, Guzmania hybrid, Heterosigma akashiwo, Hevea, Hordeum vulgare, Hypnum cupressiforme, Jatropha curcas, Karenia brevis (C.C.Davis) s) G.Hansen & Ø.Moestrup (Wilson isolate), Kochia prostrata, Lathyrus sativus, Liquidambar formosana, Malus domestica, Medicago truncatula, Micromonas pusila, Nicotiana benthamiana, Nicotiana tabacum, Panicum virgatum, Petunia hybrida cv. Mitchell, Phaeodactylum tricornutum, Physcomitrium patens, Pisum sativum, olytrichum formosum, Porosira glacialis,, Porphyra sp., Ricinus communis, Robinia pseudoacacia, Rhytidiadelphus squarrosus, Saccharum sp., Schima superba, Skeletonema costatum (diatom), Skeletonema marinoi (diatom), Solanum lycopersicum, Spinacia oleracea, lichens, Stanleya pinnata, Symbiodinium sp., Synechococcus PCC 7942, Synechococcus elongatus UTEX 2973, Rhoeo discolor, Thalassiosira pseudonana, Thermosynechococcus elongatus, Triticum aestivum, Prochlorococcus sp. (surface and deep water ecotype), Triticum aestivum, dinoflagellate endosymbionts (genus Symbiodinium), extreme acidophilic verrucomicrobial methanotroph Methylacidiphilum fumariolicum strain SolV, Thalassiosira punctigera, Tisochrysis lutea, Verbascum lychnitis, Vitis vinifera, Quercus ilex
Predicted reactivity: Alpha proteobacteria, Algae (brown and red) including Galdieria sulphuraria, Arthrospira platensis, Dicots, Benincasa hispida, Kalanchoe fedtschenkoi; Beta-proteobacteria, Conifers, Cryptomonads, Cyanobacteria (prochlorophytes), Eragrostis tef, Gamma-proeobacteria, Liverworts, Manihot esculenta, Marchantia polymorpha, Miscanthus giganteus, Monocots, Mosses, Suaeda glauca, Welwitschia; Nannochloropsis sp., Picochlorum sp., Porphyridium purpureum, Zea mays, Zosteria marina
For detection in Rhodospirillaceae use product AS15 2955
Species of your interest not listed? Contact usNot reactive in: No confirmed exceptions from predicted reactivity are currently known - Application Examples
-
Western blot 
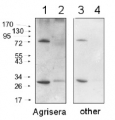
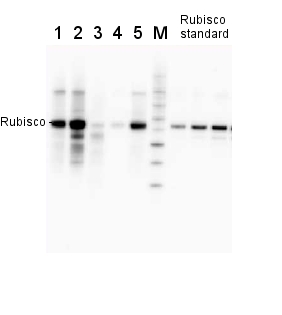
0.25 µg of chlorophyl a/lane from Spinacia oleracea (1), Synechococcus PCC 7942 (2), Cyanophora paradoxa (3), Heterosigma akashiwo (4), Thalassiosira pseudonana (5), Euglena gracilis (6), Micromonas pusilla (7), Chlamydomonas reinhardtii (8), Porphyra sp (9), Gonyaulax polyedra (10), Emiliania huxleyi (11) extracted with PEB (AS08 300), were separated on 4-12% NuPage (Invitrogen) LDS-PAGE and blotted 1h to nitrocellulose. Filters were blocked 1h with 2% low-fat milk powder in TBS-T (0.1% TWEEN 20) and probed with anti-RbcL antibody (AS03 037, 1:50 000, 1h) and secondary anti-rabbit (1:20000, 1 h) antibody (HRP conjugated, recommended secondary antibody AS09 602) in TBS-T containing 2% low fat milk powder. Antibody incubations were followed by washings in TBS-T. All steps were performed at RT with agitation. Blots were developed for 5 min with ECL Advance detection reagent according the manufacturers instructions (GE Healthcare). Images of the blots were obtained using a CCD imager (FluorSMax, Bio-Rad) and Quantity One software (Bio-Rad).
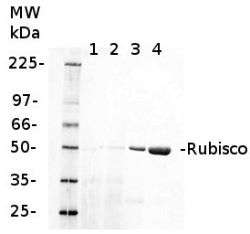
1 µg of chlorophyll from Cryptophyte samples (1,2) and 1 µg of chlorophyll (3) or 10 µg of total protein (4) from Arabidopsis thaliana leaves extracted either with 2 ml of 100 mM TrisHCl, 50 mM EDTA, 250 mM NaCl, 0.05% SDS (Sample 1) or 10 mL of 50 mM Hepes-KOH (pH 7.8), 330 mM sorbitol, 10 m EDTA, 5 mM NaCl, 5 mM MgCl2, 5 mM sodium ascorbate and 0.2% BSA (Sample 2). Samples were denatured with 1:1 Amersham WB Loading Bufferv at 70°C for 10 min and were separated on pre-casted 13.5% Amersham WB gel and blotted for 30 min to Amersham WB PVDF using wet transfer. Blots were blocked with 2% Amersham ECL Blocking Agent for 1h at room temperature (RT) with agitation. Blot was incubated in the primary antibody at a dilution of 1: 10 000 (rabbit anti-Rubisco AS03 037) for 1.5 h at RT with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, then washed once for 15 min and 3 times for 5 min in TBS-T at RT with agitation. Membrane was cut in half and left part was incubated in anti-rabbit DyLight® 550 secondary antibody from Agrisera (AS11 1782) diluted to 1:2 000 in TBST for 1h at RT with agitation. The blot was scanned using Cy3 channel of Amersham WB System.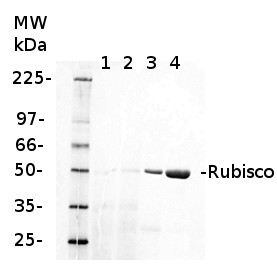
Courtesy of Dr. Małgorzata Wessels, Agrisera
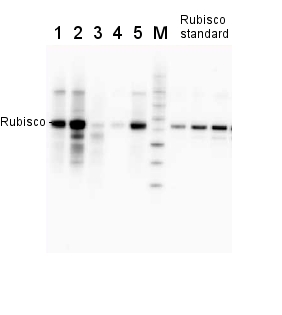
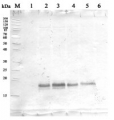
2 µg of total protein from various plant extracts (1-5) extracted with PEB (AS08 300) separated on 4-12% NuPage (Invitrogen) LDS-PAGE and blotted 1h to PVDF. Markers MagicMarks (Invitrogen) (M) and Rubisco protein standard (AS01 017S) at 0.0625 pmol, 0.125 pmol, 0.25 pmol.Following standard western blot procedure this image has been obtained using a CCD imager (FluorSMax, Bio-Rad) and Quantity One software (Bio-Rad). The contour tool of the software is used to the area for quantitation and the values are background subtracted to give an adjusted volume in counts for each standard and sample.
Note: Optimal quantitation is achieved using moderate sample loads per gel lane, generally 0.5 to 2.5 ug total protein, depending on the abundance of the target protein.
Application examples: 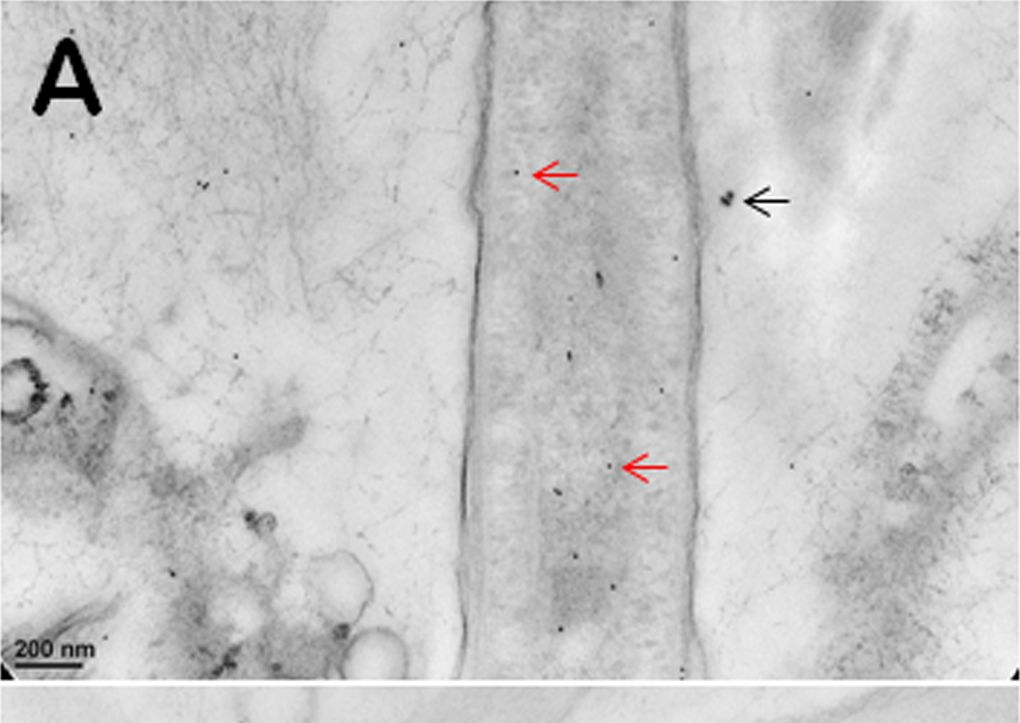
Reactant: Cyanobacteria
Application: Immunocytochemistry
Pudmed ID: 24516596
Journal: PLoS One
Figure Number: 2A
Published Date: 2014-02-12
First Author: Vali, H.
Impact Factor: 2.942
Open PublicationImmunogold TEM images of cyanobacterial cells from YNP samples.All cells were immunogold labelled by Rabbit anti-RuBisCO (large subunit, form I and II, AS03 037) at a dilution of 1000X. The gold particles are uniformly sized and dark, two examples are marked by red arrows in (A, B, C). (A) The distribution of immunogold particles tagging free RuBisCO in a cell lacking cyanophycin or carboxysomes. SiO2 around the cell is marked by a black arrow, these granules are bigger than gold particles. (B) The absence of immunogold particles in cyanophycin granules indicates the absence of RuBisCO. (C) Immunogold particles tagging RuBisCO in small inclusions suggest the presence of morphologically atypical carboxysomes. These inclusions are rare and are found in the same areas where large cyanophycin granules typically occur. Few gold particles outside the cell in (A, B, C) indicate a low level backgroung. Scale bars: 200 nm in (A, B), and 100 nm in (C).
Reactant: Cyanobacteria
Application: Western Blotting
Pudmed ID: 21445264
Journal: PLoS One
Figure Number: 5B
Published Date: 2011-03-30
First Author: Zilliges, Y., Kehr, J. C., et al.
Impact Factor: 2.942
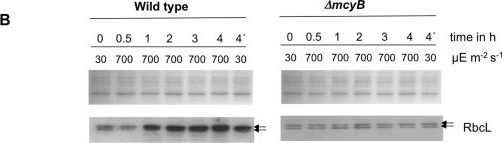
Open PublicationQuantitative analysis of RbcL.A) Immunoblot analysis of in vitro expressed and affinity purified RbcL protein using a RbcL-specific antibody and a microcystin-specific antibody, respectively. Microcystin was added as indicated. B) Representative Coomassie-stained gel pictures (upper panel) and immunoblot analyses (lower panel) with a RbcL-specific antibody of soluble protein extracts of M. aeruginosa PCC 7806 wild type (left) and the ΔmcyB mutant (right) after high light treatment of 700 µmol photons m−2 s−1 for up to four hours. The irradiation time of the individual samples is indicated. The sample 4′h was irradiated with high light for three hours and subsequently transferred to low light for one hour.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 25835989
Journal: PLoS One
Figure Number: 2B
Published Date: 2015-04-04
First Author: Fristedt, R., Martins, N. F., et al.
Impact Factor: 2.942
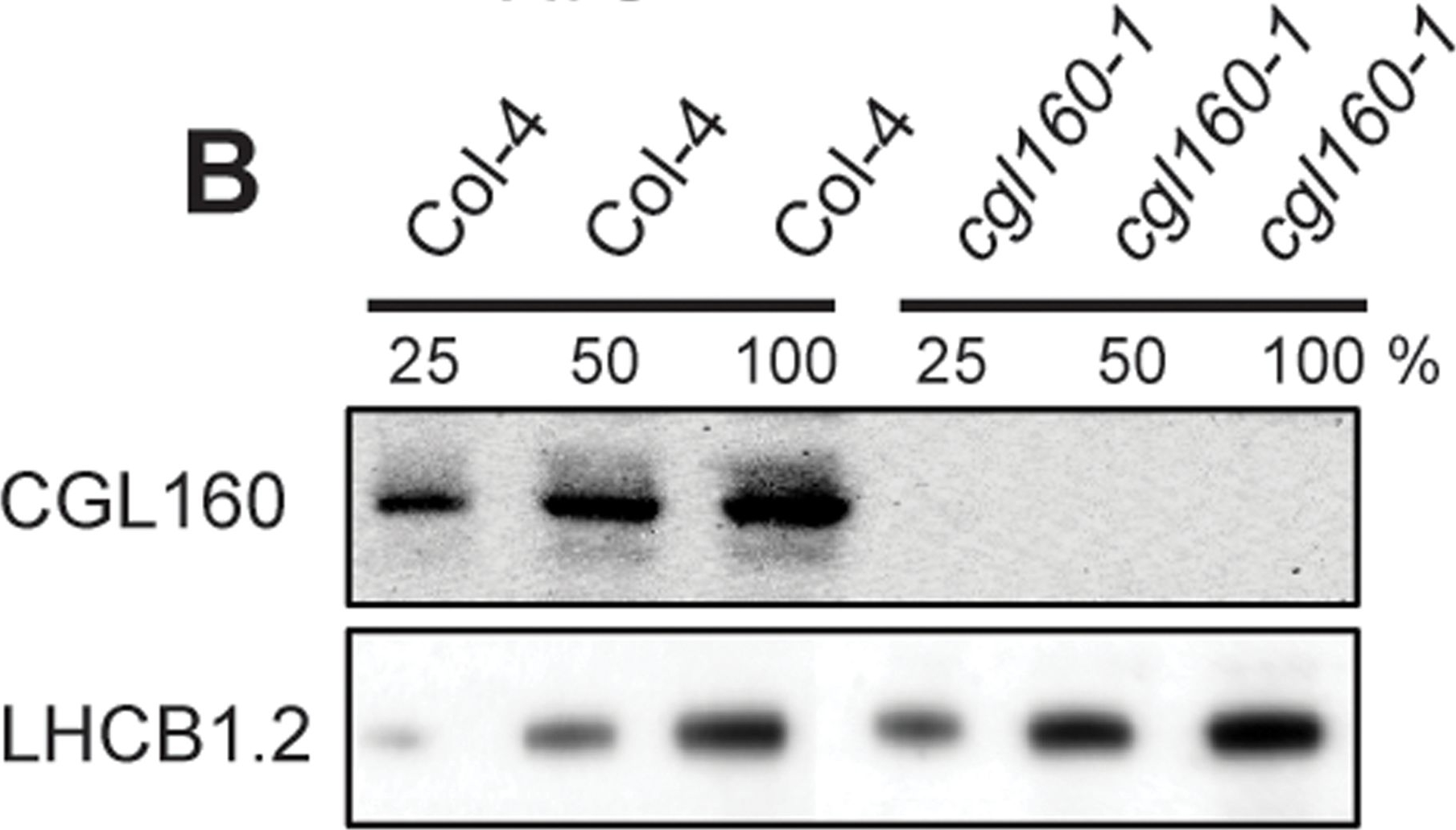
Open PublicationIsolation and characterization of Arabidopsis cgl160 mutants.A. DNA insertion sites in CGL160 gene of Arabidopsis. DNA insertion sites (black triangles) are shown in relation to the CGL160 gene structure. The two cgl160 alleles analyzed in this study are denoted as cgl160-1 and cgl160-2. The CGL160 coding region is indicated by the translational start (ATG). The CGL160 genomic locus contains nine exons but only the first four are shown in the fig (grey boxes), shown are also the first four introns (black thin connecting lines). Before ATG is the promoter region in light gray. The region used for CGL160 specific antibody is shown as antigen. B. Characterization of CGL160 amount in Arabidopsis cgl160-1 mutant from isolated chloroplasts. The CGL160 antibody was used for immunoblotting and 10 μg protein was loaded in each lane. The LHCB1 antibody was used as a loading control. C. Characterization of CGL160 amount in Arabidopsis cgl160-2 mutant from isolated chloroplasts. The CGL160 antibody was used for immunoblotting and 10 μg protein was loaded in each lane. The LHCB1 antibody was used as a loading control.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 25835989
Journal: PLoS One
Figure Number: 2C
Published Date: 2015-04-04
First Author: Fristedt, R., Martins, N. F., et al.
Impact Factor: 2.942
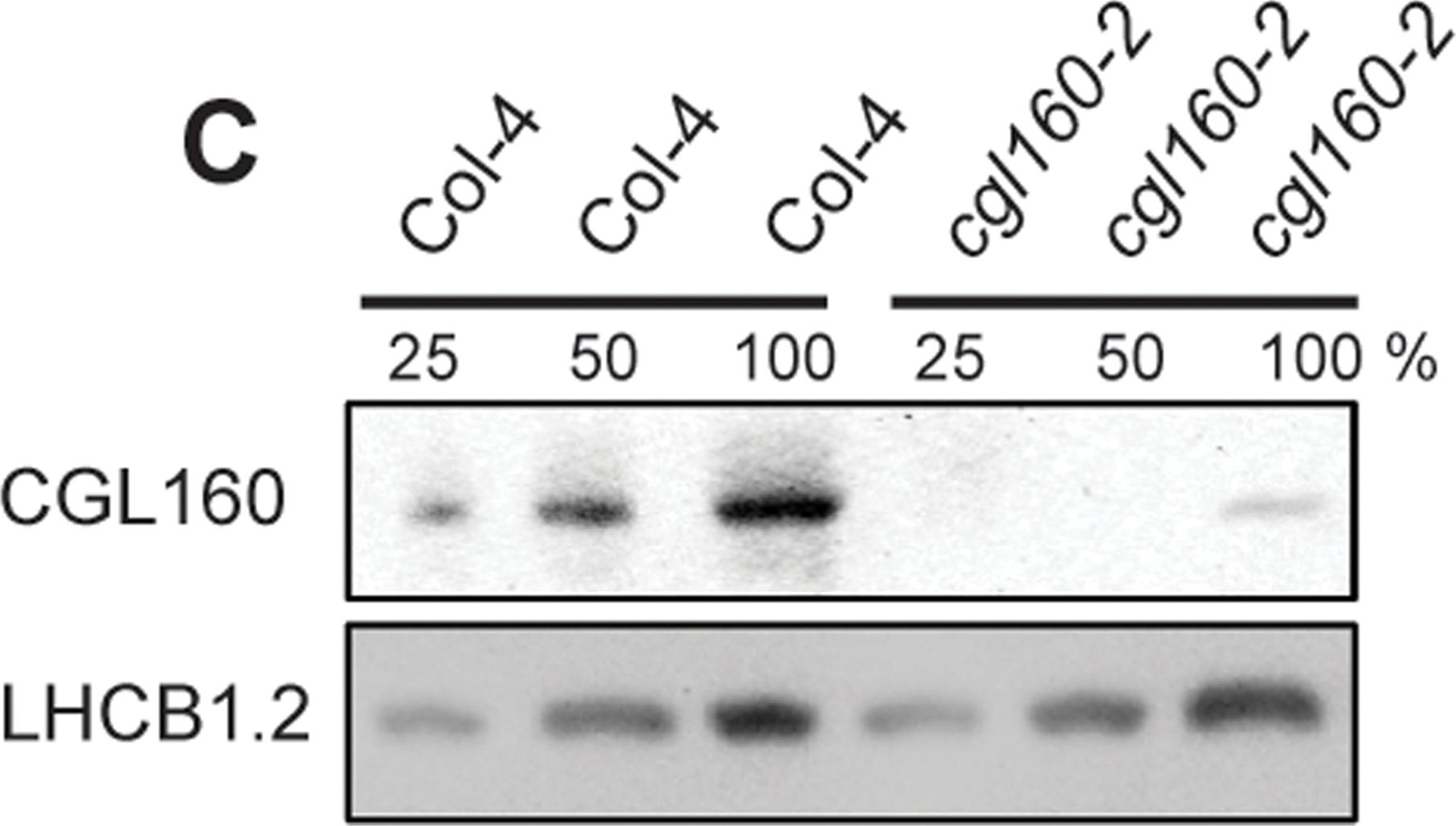
Open PublicationIsolation and characterization of Arabidopsis cgl160 mutants.A. DNA insertion sites in CGL160 gene of Arabidopsis. DNA insertion sites (black triangles) are shown in relation to the CGL160 gene structure. The two cgl160 alleles analyzed in this study are denoted as cgl160-1 and cgl160-2. The CGL160 coding region is indicated by the translational start (ATG). The CGL160 genomic locus contains nine exons but only the first four are shown in the fig (grey boxes), shown are also the first four introns (black thin connecting lines). Before ATG is the promoter region in light gray. The region used for CGL160 specific antibody is shown as antigen. B. Characterization of CGL160 amount in Arabidopsis cgl160-1 mutant from isolated chloroplasts. The CGL160 antibody was used for immunoblotting and 10 μg protein was loaded in each lane. The LHCB1 antibody was used as a loading control. C. Characterization of CGL160 amount in Arabidopsis cgl160-2 mutant from isolated chloroplasts. The CGL160 antibody was used for immunoblotting and 10 μg protein was loaded in each lane. The LHCB1 antibody was used as a loading control.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 25835989
Journal: PLoS One
Figure Number: 3B
Published Date: 2015-04-04
First Author: Fristedt, R., Martins, N. F., et al.
Impact Factor: 2.942
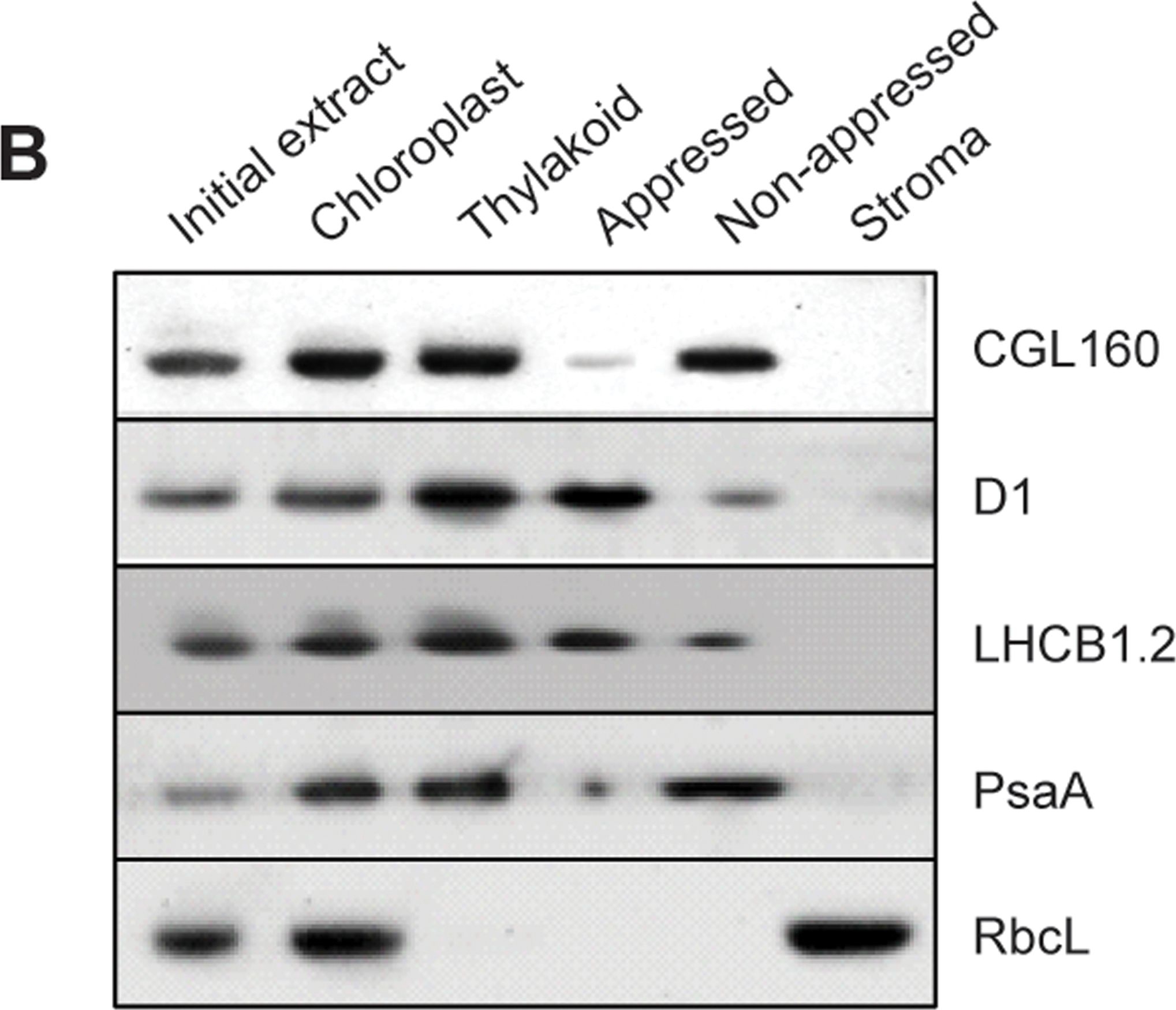
Open PublicationCGL160 is a membrane-integral chloroplast protein located in non-appressed thylakoid membranes.A. Subcellular localization of CGL160. Immunoblot analysis performed on initial plant extract, isolated chloroplasts or isolated mitochondria. TOM40 is a marker for the mitochondrion. B. Suborganellar localization determined by immunoblot analysis of chloroplast proteins diagnostic for photosystem I (PsaA), photosystem II (D1), Rubisco, LHCII (LHCB1.2) and CGL160. Protein extracts were separated by SDS-PAGE and probed with specific antibodies directed against PsaA, (PSI reaction center subunit), D1 (PSII reaction center subunits), LHCB1 (outer PSII antenna protein), the large subunit of Rubisco, a soluble protein in the stroma, and CGL160. C. Immunoblot analysis show that CG160 is an integral membrane protein associated with the thylakoid membranes. Isolated thylakoid membranes were washed with 0.4 M NaCl, and the thylakoid membranes and the supernatant were probed by immunoblotting with antibodies against CGL160 and the PSII reaction center protein D1.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 25835989
Journal: PLoS One
Figure Number: 7B
Published Date: 2015-04-04
First Author: Fristedt, R., Martins, N. F., et al.
Impact Factor: 2.942
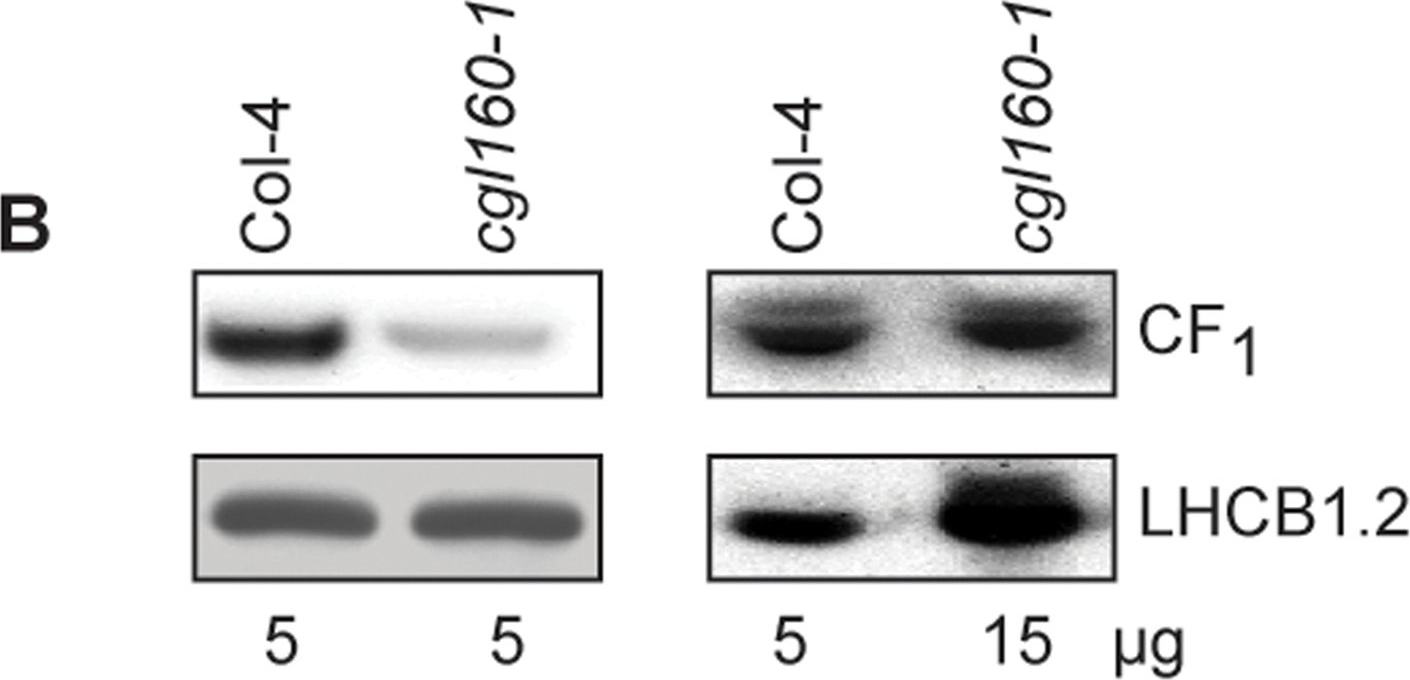
Open PublicationAltered protein accumulation and stability of the chloroplast ATP synthase in the cgl160 mutant visualized by immunoblotting.A. Immunoblots with antibodies against essential subunits of the photosynthetic protein complexes of wild-type (Col-4) Arabidopsis and the two cgl160 T-DNA insertion lines grown under long-day and short-day conditions. Isolated thylakoid membranes were used, and equal amounts of chlorophyll were loaded onto the SDS-PAGE gel. For approximate quantification, wild-type samples from long-day plants were diluted to 10%, 25% and 50%, respectively. Accumulation of PSII was probed with antibodies against PsbB and PSBO. Additionally, the PSBS protein involved in NPQ and the minor PSII antenna protein LHCB4 were probed. Accumulation of the cytochrome b6f complex was probed with antibodies against the essential subunits PetA (cytochrome f), PetB (cytochrome b6), and PETC (Rieske protein). Accumulation of PSI was probed with antibodies against the reaction center subunit PsaB and the stromal ridge subunit PsaD. ATP synthase accumulation was probed with antibodies against the CF1 subunits AtpA (CF1α), AtpB (CF1β) and AtpD (CF1δ) and antibodies against the CF0 subunits AtpF (CF0b) and AtpI (CF0a). B. Loading difference estimation for immunoblotting CF1 between wild type and cgl160-1. To obtain similar immunoblotting signal three times more (15 μg protein) was needed for cgl160-1 compared to wild type (5 μg protein). C. Maintenance of CF1 was measured by incubating leaves from wild type and cgl160-1 in solution containing the plastid protein synthesis inhibitor chloramphenicol for the indicated time points. Protein extract was isolated and separated by SDS-PAGE, immunoblotted and probed with specific antibodies against CF1 and LHCB2.1. Three times more protein was loaded from the mutant to obtain equal level of CF1 immunoblotting signal, as specified in B.
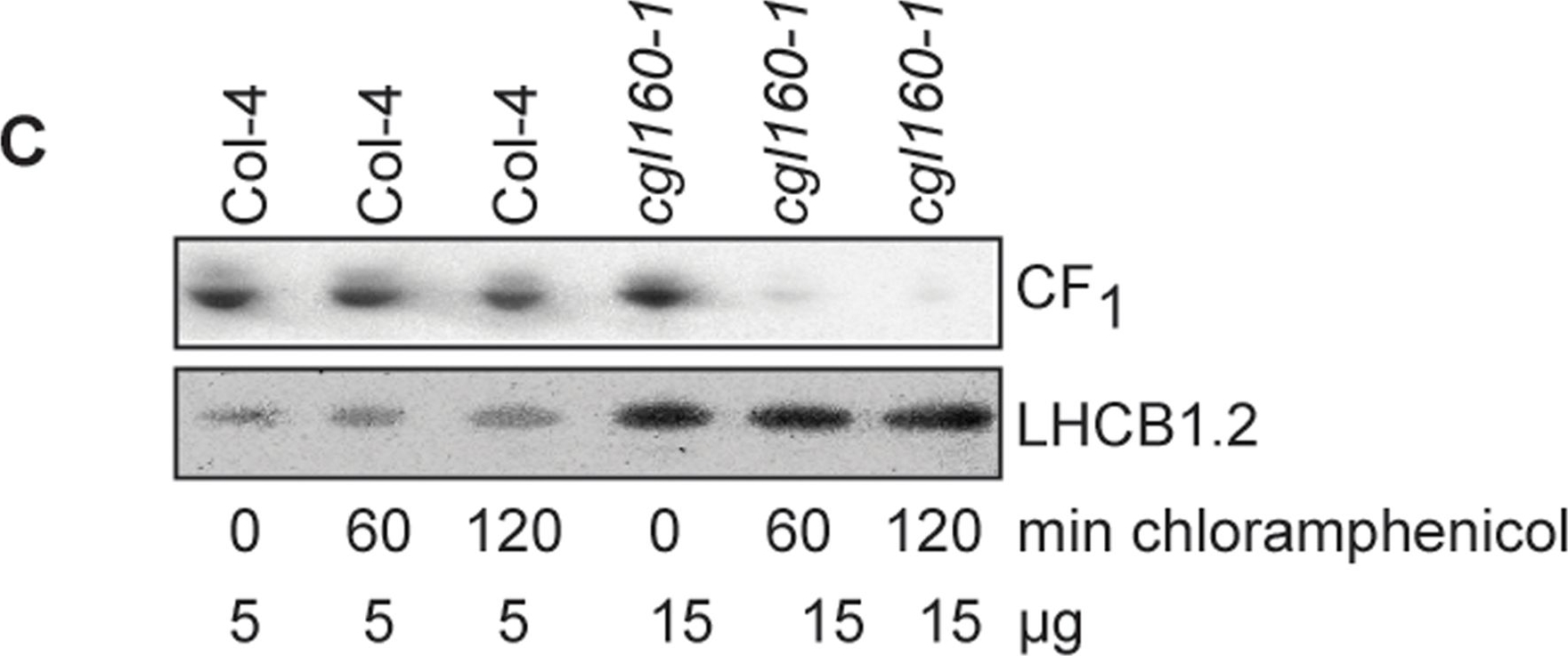
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 25835989
Journal: PLoS One
Figure Number: 7C
Published Date: 2015-04-04
First Author: Fristedt, R., Martins, N. F., et al.
Impact Factor: 2.942
Open PublicationAltered protein accumulation and stability of the chloroplast ATP synthase in the cgl160 mutant visualized by immunoblotting.A. Immunoblots with antibodies against essential subunits of the photosynthetic protein complexes of wild-type (Col-4) Arabidopsis and the two cgl160 T-DNA insertion lines grown under long-day and short-day conditions. Isolated thylakoid membranes were used, and equal amounts of chlorophyll were loaded onto the SDS-PAGE gel. For approximate quantification, wild-type samples from long-day plants were diluted to 10%, 25% and 50%, respectively. Accumulation of PSII was probed with antibodies against PsbB and PSBO. Additionally, the PSBS protein involved in NPQ and the minor PSII antenna protein LHCB4 were probed. Accumulation of the cytochrome b6f complex was probed with antibodies against the essential subunits PetA (cytochrome f), PetB (cytochrome b6), and PETC (Rieske protein). Accumulation of PSI was probed with antibodies against the reaction center subunit PsaB and the stromal ridge subunit PsaD. ATP synthase accumulation was probed with antibodies against the CF1 subunits AtpA (CF1α), AtpB (CF1β) and AtpD (CF1δ) and antibodies against the CF0 subunits AtpF (CF0b) and AtpI (CF0a). B. Loading difference estimation for immunoblotting CF1 between wild type and cgl160-1. To obtain similar immunoblotting signal three times more (15 μg protein) was needed for cgl160-1 compared to wild type (5 μg protein). C. Maintenance of CF1 was measured by incubating leaves from wild type and cgl160-1 in solution containing the plastid protein synthesis inhibitor chloramphenicol for the indicated time points. Protein extract was isolated and separated by SDS-PAGE, immunoblotted and probed with specific antibodies against CF1 and LHCB2.1. Three times more protein was loaded from the mutant to obtain equal level of CF1 immunoblotting signal, as specified in B.
Reactant: Plant
Application: Western Blotting
Pudmed ID: 27590049
Journal: BMC Plant Biol
Figure Number: 9A
Published Date: 2016-09-02
First Author: Mazur, R., Sadowska, M., et al.
Impact Factor: 4.142
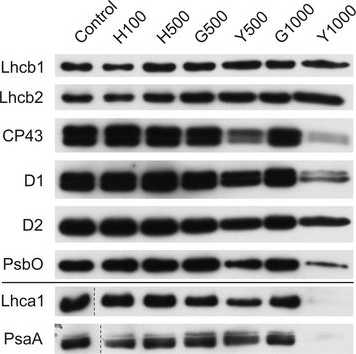
Open PublicationChanges of PSII and PSI antenna and core protein levels. Proteins from control and Tl-treated white mustard leaves were separated by SDS-PAGE followed by immunodetection with antibodies against Lhcb1, Lhcb2, Lhca1 (antenna proteins) and D1, D2, CP43, PsbO, PsaA (core proteins). Samples were loaded on the equal amount of chlorophyll (0.25 μg). Description of samples abbreviation as given in the legend to Fig. 3
Reactant: Hordeum vulgare (Barley)
Application: Western Blotting
Pudmed ID: 28338757
Journal: J Exp Bot
Figure Number: 6A
Published Date: 2017-02-01
First Author: Kucharewicz, W., Distelfeld, A., et al.
Impact Factor: 6.088
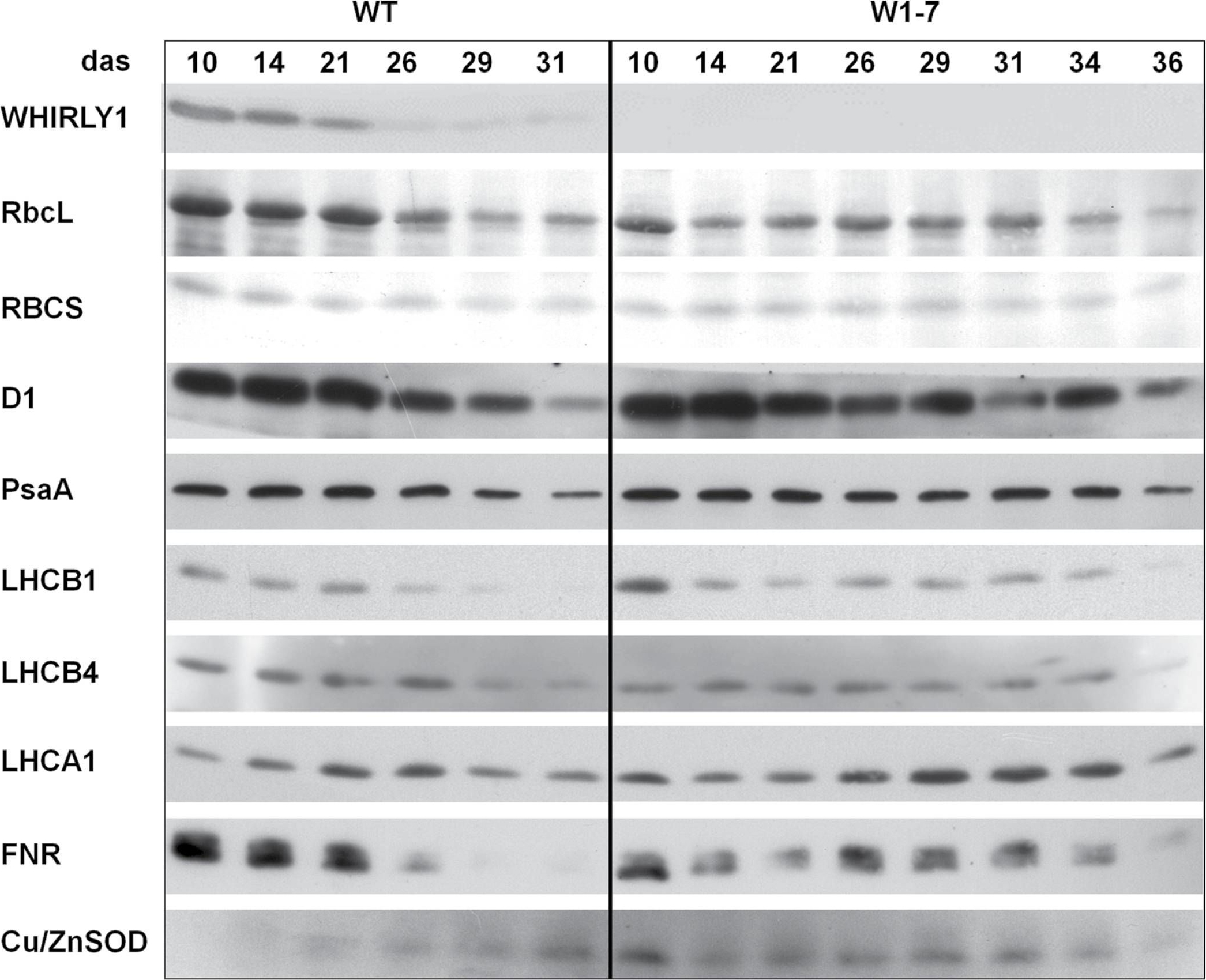
Open PublicationImmunological analyses of the relative amounts of photosynthesis-associated proteins during senescence of primary leaves from wild-type (WT) and RNAi-W1-7 plants grown at high irradiance. Samples were prepared from wild-type plants grown for 10, 14, 21, 26, 29, and 31 das, as well as from RNAi-W1-7 plants grown for 10, 14, 21, 24, 29, 31, 34, and 36 das. The analysis was done with two biological replicates. WHIRLY1, photosynthesis-related proteins, and Cu/ZnSOD were detected by specific antibodies.
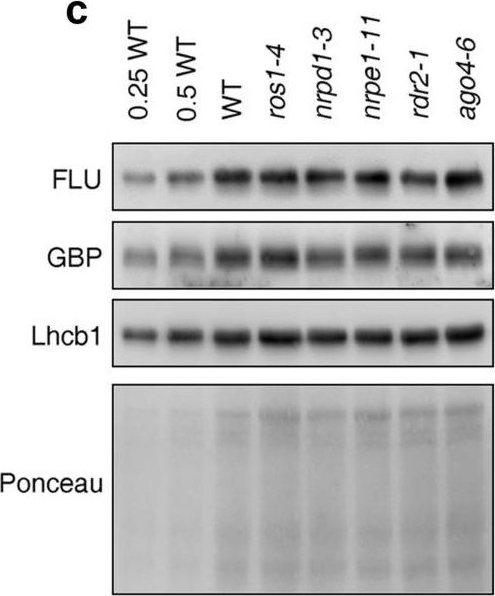
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 32963291
Journal: Sci Rep
Figure Number: 2C
Published Date: 2020-09-22
First Author: Wang, L., Leister, D., et al.
Impact Factor: 4.13
Open PublicationPerturbation of the RdDM pathway does not significantly affect chlorophyll biosynthesis. (a) Determination of total chlorophyll (Chl a + b) contents of 4-day-old seedlings grown under LD conditions. Chlorophyll was acetone-extracted and measured spectrophotometrically, and concentrations were determined as described (see “Methods”). Data are shown as mean values ± SD from 6 different plant pools. Each pool contained more than 100 seedlings. Significant differences (t-test; p < 0.05) with respect to Col-0 are indicated by asterisks. (b) Graph displaying the Chl a/b ratio of chlorophylls extracted in (a). (c) Immunoblot analysis of representative enzymes involved in chlorophyll biosynthesis (FLU and GBP) and the light-harvesting chlorophyll a/b binding protein Lhcb1. Total protein extracts from 4-day-old seedlings obtained from the wild-type (Col-0) and representative (de)methylation mutants were fractionated by SDS-PAGE, and blots were probed with antibodies raised against the individual proteins. Increasing levels of wild-type proteins were loaded in the lanes marked 0.25 WT, 0.5 WT and WT. The relative loading amounts of each sample were visualized by staining the blot with Ponceau S. FLU, FLUORESCENT IN BLUE LIGHT; GBP, glutamyl-tRNA reductase-binding protein. The figure was assembled from different blots (delineated by a black rectangle) and full-length blots are presented in Supplementary Fig. S6.
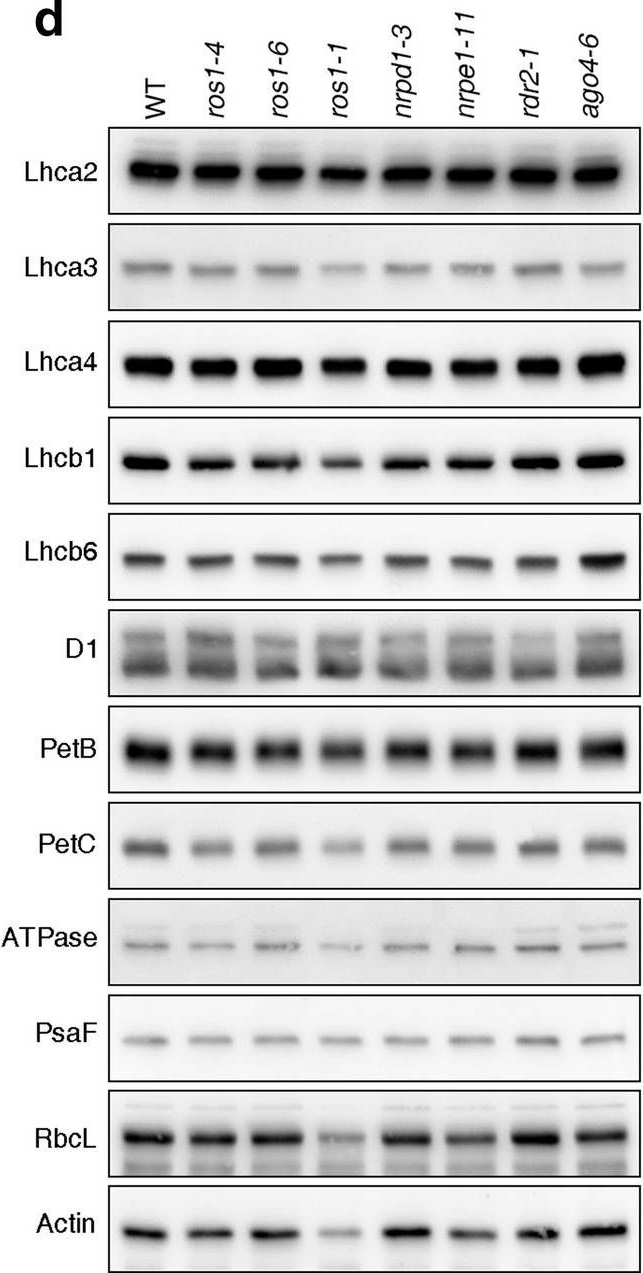
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 32963291
Journal: Sci Rep
Figure Number: 3D
Published Date: 2020-09-22
First Author: Wang, L., Leister, D., et al.
Impact Factor: 4.13
Open PublicationPhotomorphogenesis is not significantly affected in ros1, nrpd1, nrpe1, rdr2 or ago4 mutant seedlings. (a) Phenotypes (upper panel) and the corresponding maximum quantum yields of PSII (Fv/Fm) (lower panel) of 4-day-old etiolated seedlings. Fv/Fm was measured with an imaging Chl fluorometer (Imaging PAM). Scale bar = 1 cm. (b) Phenotypes (upper panel) and corresponding Fv/Fm values (lower panel) of 3-day-old etiolated seedlings which had been exposed to continuous light for 24 h. (c) Immunoblot analysis of the PSII core proteins (D1 and D2), Lhcb1 and FLU during greening of etiolated seedlings. WT seedlings were grown for 3 days in the dark and exposed to light for between 0 and 48 h, as indicated. Extracted total proteins were normalized with respect to fresh weight and fractionated by SDS-PAGE. Blots were then probed with antibodies raised against the individual proteins. Total proteins from 5-day-old WT seedlings grown under continuous light (LL) and LD conditions (LD) were used as positive controls. The total protein accumulation of each sample was visualized by staining the gel with Coomassie Blue R250 (C.B.). The figure was assembled from different blots (delineated by a black rectangle) and full-length blots are presented in Supplementary Fig. S7. (d) Immunoblot analysis of representative photosynthesis proteins of 3-day-old etiolated mutant seedlings which had been exposed to continuous light for 24 h. Immunoblot analysis was performed as in (c). The figure was assembled from different blots (delineated by a black rectangle) and full-length blots are presented in Supplementary Fig. S8. (e) Real-time PCR analyses of 3-day-old etiolated WT (Col-0) and mutant seedlings that had been exposed to continuous light for 24 h. Real-time PCR was performed with primers specific for the nuclear genes LHCB1.2, LHCB2.1, LHCB6, LHCA5, PSBP-1 and PSBTn, and the plastid genes psaA and atpB. Note that the primers for LHCB2.1 also amplify LHCB2.2 mRNA. Expression values are reported relative to the corresponding transcript levels in the WT and were normalized with respect to the expression level of ACTIN2. Data are shown as mean values ± SD from three different plant pools.
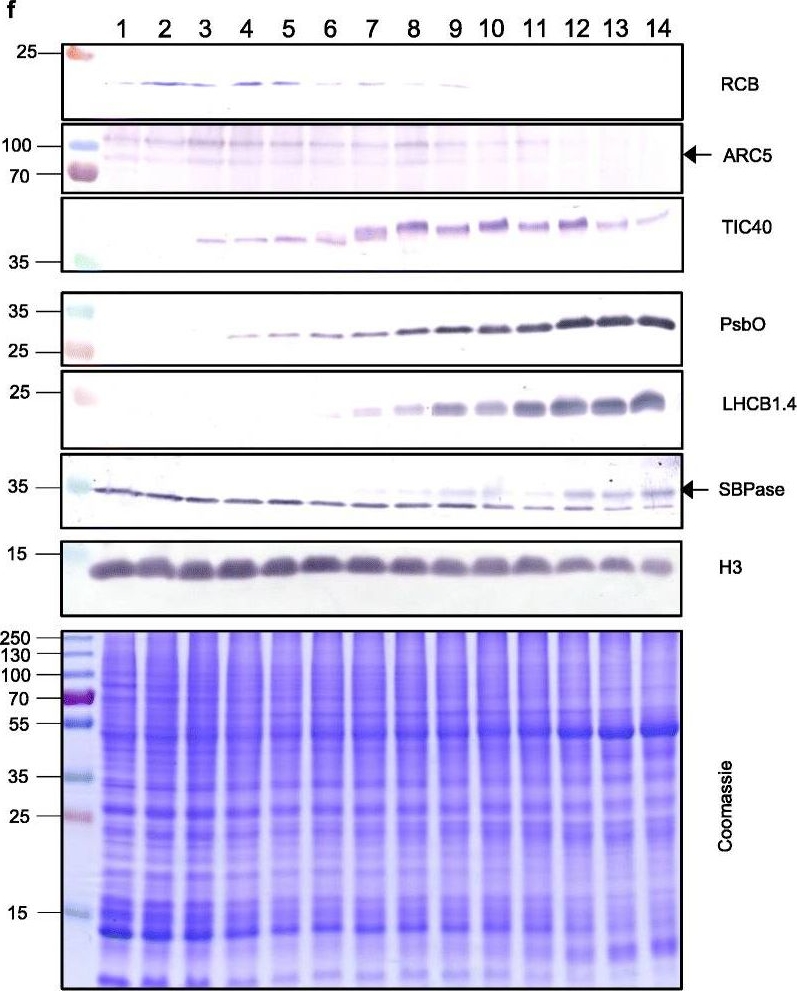
Reactant: Triticum aestivum (Common wheat)
Application: Western Blotting
Pudmed ID: 33975629
Journal: Genome Biol
Figure Number: 5F
Published Date: 2021-05-11
First Author: Loudya, N., Mishra, P., et al.
Impact Factor: 13.214
Open PublicationThe second, chloroplast growth phase involves greening and is supported by protein accumulation profiles. a Chlorophyll content, quantified in each of three independent biological replicates per sample. Error bars represent standard error of the mean. See Additional file 5: Table S6 for calculations. b, c, d Expression (Z-scores) of pigment biosynthesis and thylakoid biogenesis (b), light reactions (c) and carbon fixation-associated genes (d). e Expression (Z-scores) of chloroplast development-associated transcripts reflecting two stages of plastid development, peaking in the early plastid phase (RCB, ARC5 and TIC40) and, second, chloroplast phase (PSBO2, LHCB1.4 and SBPAse). f Immunoblot analysis of the protein products of the genes displayed in e. In total, 20 μg of protein of samples 1–14 (for PSBO2, LHCB1.4 and SBPAse), 40 μg (for RCB, ARC5 and TIC40) or 10 μg (for Histone H3 as a constitutive control) was separated on denaturing SDS-PAGE gels, transferred to blots and probed with antibodies against the protein indicated. A Coomassie-stained total protein replica gel is also shown. Molecular weights (KDa) are indicated on the left. The results show one typical example from among three independent protein extraction and immunoblot experiments
- Additional Information
-
Additional information: Anti-RbcL can be used as a cellular [compartment marker] of plastid stroma (cytoplasm in cyanobacteria) and detects RbcL protein from 31.25 fmoles. As both forms (I and II) are detected it is suitable for work with samples from Dinoflagellates, Haptophytes and Ochrophytes (diatoms, Raphidophytes, brown algae) as well as higher plants. This antibody together with Agrisera Rubisco protein standard is very suitable to quantify Rubisco in plant and algal samples.Example of a simulataneous western blot detection with RbcL, PsbA and PsaC antibodies.
This antibody is not suitable for use in immunoprecipiation.
This product can be sold containing ProClin if requested.Additional information (application): This antibody was used in:
Immunocytochemical staining of diatoms according to Schmid (2003) J Phycol 39: 139-153 and Wordemann et al. (1986) J Cell Biol 102: 1688-1698.
Immunofluorescence Dreier et al. (2012). FEMS Microbial Ecol., March 2012.
Western blot and tissue printing during a student course Ma et al. (2009).
As a loading control Sun et al. (2020).
Protocol for Rubisco quantification using this antibody can be found here. - Background
-
Background: This antibody is especially suitable for quantifying of Rubisco in plant and algal samples.
Rubisco (Ribulose-1,5-bisphosphate carboxylase/oxygenase) catalyzes the rate-limiting step of CO2 fixation in photosynthetic organisms. It is demonstrably homologous from purple bacteria to flowering plants and consists of two protein subunits, each present in 8 copies. In plants and green algae, the large subunit (~55 kDa) is coded by the chloroplast rbcL gene, and the small subunit (15 kDa) is coded by a family of nuclear rbcS genes.
- Product Citations
-
Selected references: Krupinska et al. (2025). Iron allocation to chloroplast proteins depends on the DNA-binding protein WHIRLY1. Planta. 2025 Jun 17;262(2):32. doi: 10.1007/s00425-025-04736-8.
Archer et al. (2025). Effects of chaperone selectivity on the assembly of plant Rubisco orthologs in E. coli. J Exp Bot . 2025 Mar 31:eraf140. doi: 10.1093/jxb/eraf140.
Zhou et al. (2024). Antagonistic systemin receptors integrate the activation and attenuation of systemic wound signaling in tomato. Dev Cell. 2024 Nov 26:S1534-5807(24)00670-1. doi: 10.1016/j.devcel.2024.11.005.
Pica et al. (2024). Functional ecological traits in young and adult thalli of canopy-forming brown macroalga Gongolaria barbata (Phaeophyta) from a transitional water system. PeerJ. 2024 Sep 12:12:e17959. doi: 10.7717/peerj.17959.
Phukan et al. (2024). Externally supplied ascorbic acid moderates detrimental effects of UV-C exposure in cyanobacteria. Photochem Photobiol Sci. 2024 Jul 12. doi: 10.1007/s43630-024-00612-8.
Zhao et al. (2024). Psb28 protein is indispensable for stable accumulation of PSII core complexes in Arabidopsis.Plant J. 2024 May 26. doi: 10.1111/tpj.16844.
Nuamzanei et al. (2024). Impact of polyvinyl chloride (PVC) microplastic on growth, photosynthesis and nutrient uptake of Solanum lycopersicum L. (Tomato). Environ Pollut. 2024 Apr 16:123994. doi: 10.1016/j.envpol.2024.123994.
Ciesielska et al. (2024). S2P2-the chloroplast-located intramembrane protease and its impact on the stoichiometry and functioning of the photosynthetic apparatus of A. thaliana. Front Plant Sci. 2024 Mar 15:15:1372318. doi: 10.3389/fpls.2024.1372318.
Rodrigues et al. (2023). Are tomato plants co-exposed to heat and salinity able to ensure a proper carbon metabolism?–An insight into the photosynthetic hub. Plant Physiol Biochem. 2023 Dec 10:206:108270.doi: 10.1016/j.plaphy.2023.108270.
Rredhi et al. (2023). The UV-A Receptor CRY-DASH1 Up- and Downregulates Proteins Involved in Different Plastidial Pathways. J Mol Biol. 2023 Sep 10:168271.doi: 10.1016/j.jmb.2023.168271.
Hao and Malnoë (2023). A Simple Sonication Method to Isolate the Chloroplast Lumen in Arabidopsis thaliana.Bio Protoc. 2023 Aug 5; 13(15): e4756.
Sprecher et al (2023) Genetic transformation methods for diatom Nitzschia captiva: New tools to better understand dinotom endosymbiosis
Leister et al (2023) An ancient metabolite damage-repair system sustains photosynthesis in plants
Yamada et al (2023) Prey preference in a kleptoplastic dinoflagellate is linked to photosynthetic performance
Thabet et al. (2023) Characterization of cellular toxicity induced by sub-lethal inorganic mercury in the marine microalgae Chlorococcum dorsiventrale isolated from a metal-polluted coastal site. Chemosphere. 2023 Jul 4:139391. doi: 10.1016/j.chemosphere.2023.139391.
Cruz LP, Pacheco VS, Nogueira CCM, et al. Variation of photosynthesis along the canopy profile of sugarcane and energy canes. Research Square; 2023. DOI: 10.21203/rs.3.rs-3124093/v1
Vitale, L.; Vitale, E.; Francesca, S.; Lorenz, C.; Arena, C. Plant-Growth Promoting Microbes Change the Photosynthetic Response to Light Quality in Spinach. Plants 2023, 12, 1149. https://doi.org/10.3390/plants12051149
Koskinen et al. (2023) .The genome sequence of Synechocystis sp. PCC 6803 substrain GT-T and its implications for the evolution of PCC 6803 substrains
Aguiló-Nicolau et al. (2023) Singular adaptations in the carbon assimilation mechanism of the polyextremophile cyanobacterium Chroococcidiopsis thermalis. Photosynth Res. 2023 May;156(2):231-245. doi: 10.1007/s11120-023-01008-y.
Cano-Ramirez et al. (2023) Low-temperature and circadian signals are integrated by the sigma factor SIG5. Nat Plants. 2023 Apr;9(4):661-672. doi: 10.1038/s41477-023-01377-1. Epub 2023 Mar 30.
Boussardon C, Carrie C, Keech O. Comparing plastid proteomes points towards a higher plastidial redox turnover in vascular tissues than in mesophyll cells. J Exp Bot. 2023 Apr 7:erad133. doi: 10.1093/jxb/erad133.
Chen et al. (2023) Producing fast and active Rubisco in tobacco to enhance photosynthesis. Plant Cell. 2023;35(2):795-807. doi:10.1093/plcell/koac348
Garcia et al. (2023) Effects of RuBisCO and CO2 concentration on cyanobacterial growth and carbon isotope fractionation [published online ahead of print, 2023 Jan 5]. Geobiology. 2023;10.1111/gbi.12543. doi:10.1111/gbi.12543
Minagawa, Dann. (2023) Extracellular CahB1 from Sodaline magerasimenkoae IPPAS B-353 Acts as a Functional Carboxysomal beta-Carbonic Anhydrase in Synechocystis sp. PCC6803. Plants (Basel). 2023;12(2):265. Published 2023 Jan 6. doi:10.3390/plants12020265
Vidal-Meireles, et al. (2023)The lifetime of the oxygen-evolving complex subunit PSBO depends on light intensity and carbon availability in Chlamydomonas. Plant Cell Environ. 2023;46(2):422-439. doi:10.1111/pce.14488
Capo-Bauca et al. (2023). Carbon assimilation in upper subtidal macroalgae is determined by an inverse correlation between Rubisco carboxylation efficiency and CO2 concentrating mechanism effectiveness. New Phytol. 2023;237(6):2027-2038. doi:10.1111/nph.18623
Cao et al. (2022) Autophagic pathway contributes to low-nitrogen tolerance by optimizing nitrogen uptake and utilization in tomato. Hortic Res. 2022 Mar 23;9:uhac068. doi: 10.1093/hr/uhac068. PMID: 35669705; PMCID: PMC9164271.
Li et al. (2022) The CDC48 complex mediates ubiquitin-dependent degradation of intra-chloroplast proteins in plants. Cell Rep. 2022 Apr 12;39(2):110664. doi: 10.1016/j.celrep.2022.110664. PMID: 35417702.
Capo-Bauca et al. (2022) Correlative adaptation between Rubisco and CO2-concentrating mechanisms in seagrasses. Nat Plants. 2022 Jun;8(6):706-716. doi: 10.1038/s41477-022-01171-5. Epub 2022 Jun 20. Erratum in: Nat Plants. 2022 Jun 29;: PMID: 35729266.
Rathi et al. (2022) Suspension cell secretome of the grain legume Lathyrus sativus (grasspea) reveals roles in plant development and defense responses. Phytochemistry. 2022 Oct;202:113296. doi: 10.1016/j.phytochem.2022.113296. Epub 2022 Jul 20. PMID: 35868566.
Neusius et al. (2022) Lysine acetylation regulates moonlighting activity of the E2 subunit of the chloroplast pyruvate dehydrogenase complex in Chlamydomonas. Plant J. 2022 Sep;111(6):1780-1800. doi: 10.1111/tpj.15924. Epub 2022 Aug 8. PMID: 35899410.
Takeshi Cerritos-Castro et a. (2022) Amaranth calcium oxalate crystals are associated with chloroplast structures and proteins, Microscopy Research and Technique, Volume85, Issue11, November 2022, Pages 3694-3706.
Vitale et al.(2022) Manipulation of light quality is an effective tool to regulate photosynthetic capacity and fruit antioxidant properties of Solanum lycopersicum L. cv. 'Microtom' in a controlled environment. PeerJ. 2022;10:e13677. Published 2022 Jul 1. doi:10.7717/peerj.13677
Zang, Xu, Yan & Wu (2022). Elevated CO2 modulates the physiological responses of Thalassiosira pseudonana to ultraviolet radiation [published online ahead of print, 2022 Sep 22]. J Photochem Photobiol B. 2022;236:112572. doi:10.1016/j.jphotobiol.2022.112574
Perera-Castro et al (2022). Limitations to photosynthesis in bryophytes: certainties and uncertainties regarding methodology. J Exp Bot. 2022;73(13):4592-4604. doi:10.1093/jxb/erac190
Cabezas-Fuster et al. (2022). Missplicing suppressor alleles of Arabidopsis PRE-MRNA PROCESSING FACTOR 8 increase splicing fidelity by reducing the use of novel splice sites. Nucleic Acids Res. 2022 Jun 10;50(10):5513-5527. doi: 10.1093/nar/gkac338. PMID: 35639749; PMCID: PMC9177961.
Burlacot et al. (2022) Alternative photosynthesis pathways drive the algal CO2-concentrating mechanism. Nature 605, 366–371 (2022).
Bychkov et al. (2022) The role of PAP4/FSD3 and PAP9/FSD2 in heat stress responses of chloroplast genes. Plant Sci. 2022 Sep;322:111359. doi: 10.1016/j.plantsci.2022.111359. Epub 2022 Jun 20. PMID: 35738478.
Rathi et al.(2022) Dissection of grasspea (Lathyrus sativus L.) root exoproteome reveals critical insights and novel proteins,Plant Science, Volume 316, 2022, 111161, ISSN 0168-9452.
Kumar et al. (2022). Proteomic dissection of rice cytoskeleton reveals the dominance of microtubule and microfilament proteins, and novel components in the cytoskeleton-bound polysome, Plant Physiology and Biochemistry, Volume 170,2022,Pages 75-86,ISSN 0981-9428.
Chen et al. (2022) ACS Synth. Biol. 2022, 11, 1, 154–161Publication Date:October 19, 2021
Rakhmankulova et al. (2021) Possible Activation of С3 Photosynthesis in С4 Halophyte Kochia prostrata Exposed to an Elevated Concentration of СО2. Russ J Plant Physiol 68, 1107–1114 (2021).
Mazur et al. (2021) The SnRK2.10 kinase mitigates the adverse effects of salinity by protecting photosynthetic machinery. Plant Physiol. 2021 Dec 4;187(4):2785-2802. doi: 10.1093/plphys/kiab438. PMID: 34632500; PMCID: PMC8644180.
Buck et al. (2021) Identification of sequence motifs in Lhcx proteins that confer qE-based photoprotection in the diatom Phaeodactylum tricornutum. Plant J. 2021 Dec;108(6):1721-1734. doi: 10.1111/tpj.15539. Epub 2021 Nov 3. PMID: 34651379.
Pavlovic & Kocab. (2021) Alternative oxidase (AOX) in the carnivorous pitcher plants of the genus Nepenthes: what is it good for? Ann Bot. 2021 Dec 18:mcab151. doi: 10.1093/aob/mcab151. Epub ahead of print. PMID: 34922341.
Medina-Puche et al (2021). Protocol for evaluating protein relocalization from the plasma membrane to chloroplasts. STAR Protoc. 2021 Sep 14;2(4):100816. doi: 10.1016/j.xpro.2021.100816. PMID: 34585156; PMCID: PMC8450296.
Buck et al. (2021) Identification of sequence motifs in Lhcx proteins that confer qE-based photoprotection in the diatom Phaeodactylum tricornutum. Plant J. 2021 Dec;108(6):1721-1734. doi: 10.1111/tpj.15539. Epub 2021 Nov 3. PMID: 34651379.
Kedzior et al (2021) Quantification of RuBisCO Expression and Photosynthetic Oxygen Evolution in Cyanobacteria. Bio Protoc. 2021 Oct 20;11(20):e4199. doi: 10.21769/BioProtoc.4199. PMID: 34761071; PMCID: PMC8554809.
Vitale et al. (2021) Light Spectral Composition Influences Structural and Eco-Physiological Traits of Solanum lycopersicum L. cv. 'Microtom' in Response to High-LET Ionizing Radiation. Plants (Basel). 2021 Aug 23;10(8):1752. doi: 10.3390/plants10081752. PMID: 34451797; PMCID: PMC8399554.
Cecchin et al (2021) LPA2 protein is involved in photosystem II assembly in Chlamydomonas reinhardtii. Plant J. 2021 Jul 4. doi: 10.1111/tpj.15405. Epub ahead of print. PMID: 34218480.
Bernau et al. (2021) Precision analysis for the determination of steric mass action parameters using eight tobacco host cell proteins,Journal of Chromatography A,Volume 1652,2021,462379,ISSN 0021-9673,https://doi.org/10.1016/j.chroma.2021.462379.
Cary et al. (2021) Field trial demonstrating phytoremediation of the military explosive RDX by XplA/XplB-expressing switchgrass. Nat Biotechnol. 2021 May 3. doi: 10.1038/s41587-021-00909-4. Epub ahead of print. PMID: 33941930.
Curien et al. (2021) Mixotrophic growth of the extremophile galdieria sulphuraria reveals the flexibility of its carbon assimilation metabolism. New Phytol. 2021 Mar 25. doi: 10.1111/nph.17359. Epub ahead of print. PMID: 33764540.
Sun et al. (2021). A molecular switch in sulfur metabolism to reduce arsenic and enrich selenium in rice grain. Nat Commun. 2021 Mar 2;12(1):1392. doi: 10.1038/s41467-021-21282-5. PMID: 33654102; PMCID: PMC7925690. (Loading Control)
Li et al. (2021) Isolation and comparative proteomic analysis of mitochondria from the pulp of ripening citrus fruit. Hortic Res. 2021 Feb 1;8(1):31. doi: 10.1038/s41438-021-00470-w. PMID: 33518707; PMCID: PMC7848011.
Mlinaric et el. (2021). Antioxidative response and photosynthetic regulatory mechanisms in common fig leaves after short‐term chilling stress. Ann Appl Biol. 2021; 1– 13.
Wang et al. (2020). Effects and Mechanisms of Foliar Application of Silicon and Selenium Composite Sols on Diminishing Cadmium and Lead Translocation and Affiliated Physiological and Biochemical Responses in Hybrid Rice (Oryza Sativa L.) Exposed to Cadmium and Lead. Chemosphere. 2020 Jul;251:126347.
Khajuria et al. (2020). Photochemical Efficiency Is Negatively Correlated With the Delta 9- Tetrahydrocannabinol Content in Cannabis Sativa L. Plant Physiol Biochem. 2020 Apr 8;151:589-600. doi: 10.1016/j.plaphy.2020.04.003.
Kurmayer et al. (2020). Chemically labeled toxins or bioactive peptides show a heterogeneous intracellular distribution and low spatial overlap with autofluorescence in bloom-forming cyanobacteria. Sci Rep. 2020 Feb 17;10(1):2781. doi: 10.1038/s41598-020-59381-w. (Immunofluorescence)
Zhang et al. (2020). Hydrogen sulfide and rhizobia synergistically regulate nitrogen (N) assimilation and remobilization during N deficiency-induced senescence in soybean. Plant Cell Environ. 2020 Feb 3. doi: 10.1111/pce.13736.
Zavrel et al. (2019). Quantitative insights into the cyanobacterial cell economy. Elife. 2019 Feb 4;8. pii: e42508. doi: 10.7554/eLife.42508.
Buck et al. (2019). Lhcx proteins provide photoprotection via thermal dissipation of absorbed light in the diatom Phaeodactylum tricornutum. Nat Commun. 2019 Sep 13;10(1):4167. doi: 10.1038/s41467-019-12043-6.
Saha et al. (2019). Dynamics of protein accumulation from the 3'end of viral RNA is different from the rest of the genome in potato virus A infection. J Virol. 2019 Jul 24. pii: JVI.00721-19. doi: 10.1128/JVI.00721-19. (loading control)
Schober et al. (2019). Organelle Studies and Proteome Analyses on Mitochondria and Plastids Fractions from the Diatom Thalassiosira pseudonana. Plant Cell Physiol. 2019 Jun 10. pii: pcz097. doi: 10.1093/pcp/pcz097.
Lacour et al. (2019). Decoupling light harvesting, electron transport and carbon fixation during prolonged darkness supports rapid recovery upon re-illumination in the Arctic diatom Chaetoceros neogracilis. Polar Biol (2019).
Contreras et al. (2019). UV-B shock induces photoprotective flavonoids but not antioxidant activity in Antarctic Colobanthus quitensis (Kunth) Bartl. Environmental and Experimental Botany, Volume 159, March 2019, Pages 179-190.
Deng et al. (2019). Integrated proteome analyses of wheat glume and awn reveal central drought response proteins under water deficit conditions. J Plant Physiol. 2019 Jan;232:270-283. doi: 10.1016/j.jplph.2018.11.011.
Kong et al. (2018) Interorganelle Communication: Peroxisomal MALATE DEHYDROGENASE2 Connects Lipid Catabolism to Photosynthesis through Redox Coupling in Chlamydomonas. Plant Cell. 2018 Aug;30(8):1824-1847. doi: 10.1105/tpc.18.00361
Pao et al. (2018). Lamelloplasts and minichloroplasts in Begoniaceae: iridescence and photosynthetic functioning. J Plant Res. 2018 Mar 2. doi: 10.1007/s10265-018-1020-2. (ImmunoGold)
Deng et al. (2018). Comparative Proteome Analysis of Wheat Flag Leaves and Developing Grains Under Water Deficit. Front Plant Sci. 2018 Apr 10;9:425. doi: 10.3389/fpls.2018.00425. eCollection 2018.
Ravi et al. (2018). Separation Options for Phosphorylated Osteopontin from Transgenic Microalgae Chlamydomonas reinhardtii. Int J Mol Sci. 2018 Feb 16;19(2). pii: E585. doi: 10.3390/ijms19020585.
Wu et al. (2018). Control of Retrograde Signaling by Rapid Turnover of GENOMES UNCOUPLED 1. Plant Physiol. 2018 Jan 24. pii: pp.00009.2018. doi: 10.1104/pp.18.00009.
Kim et al. (2017). Effect of cell cycle arrest on intermediate metabolism in the marine diatom Phaeodactylum tricornutum. Proc Natl Acad Sci U S A. 2017 Sep 19;114(38):E8007-E8016. doi: 10.1073/pnas.1711642114.
Arena et al. (2017). Eco-physiological and Antioxidant Responses of Holm Oak (Quercus ilex L.) Leaves to Cd and Pb. Water, Air, & Soil Pollution December 2017, 228:459.
Jespersen et al. (2017). Metabolic Effects of Acibenzolar-S-Methyl for Improving Heat or Drought Stress in Creeping Bentgrass. Front Plant Sci. 2017 Jul 11;8:1224. doi: 10.3389/fpls.2017.01224. eCollection 2017. (western blot, Agostis stolonifera cv. ‘Penncross’)
Neto et al. (2017). Cyclic electron flow, NPQ and photorespiration are crucial for the establishment of young plants of Ricinus communis and Jatropha curcas exposed to drought. Plant Biol (Stuttg). 2017 Apr 12. doi: 10.1111/plb.12573. (Jatropha curcas and Ricinus communis, western blot)
Ribeiro et al. (2017). Increased sink strength offsets the inhibitory effect of sucrose on sugarcane photosynthesis. J Plant Physiol. 2017 Jan;208:61-69. doi: 10.1016/j.jplph.2016.11.005.
Baumgart et al. (2017). Heterologous expression of the Halothiobacillus neapolitanus carboxysomal gene cluster in Corynebacterium glutamicum. J Biotechnol. 2017 Mar 27. pii: S0168-1656(17)30124-4. doi: 10.1016/j.jbiotec.2017.03.019.
Kolesinski et al. (2017). Is RAF1 protein from Synechocystis sp. PCC 6803 really needed in the cyanobacterial Rubisco assembly process. Photosynth Res. 2017 Jan 20. doi: 10.1007/s11120-017-0336-4.
Castiglia et al. (2016). High-level expression of thermostable cellulolytic enzymes in tobacco transplastomic plants and their use in hydrolysis of an industrially pretreated Arundo donax L. biomass.Biotechnol Biofuels. 2016 Jul 22;9:154. doi: 10.1186/s13068-016-0569-z. eCollection 2016.
Meng et al. (2016). Physiological and proteomic responses to salt stress in chloroplasts of diploid and tetraploid black locust (Robinia pseudoacacia L.). Sci Rep. 2016 Mar 15;6:23098. doi: 10.1038/srep23098
Heinnickel et al. (2016). Tetratricopeptide repeat protein protects photosystem I from oxidative disruption during assembly. Proc Natl Acad Sci U S A. 2016 Mar 8;113(10):2774-9. doi: 10.1073/pnas.1524040113
Young et al. (2015). Antarctic phytoplankton down-regulate their carbon-concentrating mechanisms under high CO2 with no change in growth rates. Marine Ecology Progress Series 532:13-28.
Li at al. (2015). Salt stress response of membrane proteome of sugar beet monosomic addition line M14. J Proteomics. 2015 Apr 3. pii: S1874-3919(15)00109-8. doi: 10.1016/j.jprot.2015.03.025.
Krasuska et al. (2015). Switch from heterotrophy to autotrophy of apple cotyledons depends on NO signal. Planta. 2015 Jul 18.
Janeczko et al. (2015). Disturbances in production of progesterone and their implications in plant studies. Steroids. 2015 Feb 9. pii: S0039-128X(15)00054-9. doi: 10.1016/j.steroids.2015.01.025.
Armbruster et al. (2014). Ion antiport accelerates photosynthetic acclimation in fluctuating light environments. Nat Commun. 2014 Nov 13;5:5439. doi: 10.1038/ncomms6439
Kolesinski et al. (2014). Rubisco Accumulation Factor 1 from Thermosynechococcus elongatus participates in the final stages of ribulose-1,5-bisphosphate carboxylase/oxygenase assembly in Escherichia coli cells and in vitro. FEBS J. 2014 Jul 12. doi: 10.1111/febs.12928
Pandey and Pandey-Rai (2014). Modulations of physiological responses and possible involvement of defense-related secondary metabolites in acclimation of Artemisia annua L. against short-term UV-B radiation. Planta. 2014 Jul 15.
Liang et al. (2014). Cyanophycin mediates the accumulation and storage of fixed carbon in non-heterocystous filamentous cyanobacteria from coniform mats. PLoS One. 2014 Feb 7;9(2):e88142. doi: 10.1371/journal.pone.0088142. eCollection 2014. (immunogold)
Mayfield et al. (2014). Rubisco Expression in the Dinoflagellate Symbiodinium sp. Is Influenced by Both Photoperiod and Endosymbiotic Lifestyle. Mar Biotechnol, Jan 22.
Chen, T., Hojka, M., Davey, P. et al. Engineering α-carboxysomes into plant chloroplasts to support autotrophic photosynthesis. Nat Commun 14, 2118 (2023). https://doi.org/10.1038/s41467-023-37490-0 - Protocols
-
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extracts
Agrisera Educational Posters CollectionRubisco quantitation in plant and algal samples using Agrisera anti-RbcL global antibody and Rubisco protein standard
Methodology: Plant samples are generally ground with liquid nitrogen in a mortar and pestle. The resulting powder is transferred to a plastic tube. Algal samples can be either concentrated by centrifugation or, preferably, by filtration onto glass fiber filters. Solubilization is performed in Agrisera protein extraction buffer (PEB, AS08 300) containing 0.1mg/mL PefaBloc SC (AEBSF) protease inhibitor (Roche). Disruption is most optimally obtained through flash freezing of the sample in liquid nitrogen alternated with thawing by sonication with a microtip. This process can be repeated depending on the toughness of the sample. The sample is adjusted to 50 mM dithiothreitol and heated to 70°C for 5 minutes. Samples are cooled and centrifuged briefly prior to electrophoresis.
Optimal quantitation is achieved using moderate sample loads per gel lane, generally 0.5 to 2.5 ug total protein, depending on the abundance of the target protein.
Electrophoresis and Immunoblotting: Once solubilized, the proteins can be separated electrophoretically in a number of systems. We obtain optimal results with the Invitrogen NuPAGE gel system using Bis-Tris 4-12% gradient gels. Proteins are separated in MES SDS running buffer according to the manufacturer’s recommendations at 200 V for 35 minutes. The gels are transferred to PVDF in the same apparatus, the SureLock XCell blot module, for 60 minutes at 30 V for a single gel or 80 minutes for a pair. Following transfer the blots are blocked with non-fat dry milk up to 10 % in TBS-T, for 1 h/RT with gentle agitation. The blot is incubated with primary antibody, usually at 1:25 000 to 1:50 000, for 1 h/RT (if extreme femtogram detection reagents are used) or in lower primary antibody dilution for less sensitivie reagents (mid picogram and lower).
For quantitation a relatively high primary antibody: target protein ratio gives more reliable results than immunoblots at low ratios of primary antibody:target protein.
The blot is washed extensively in TBS-T (twice briefly, once for 15 minutes and three times for five minutes). The blot is incubated with secondary antibody, for example goat anti-rabbit IgG horse radish peroxidase conjugated, AS09 602 (Agrisera), for 1h/RT. The blot is washed as above and developed with ECL detection reagents.Quantitation: When quantitated standards are included on the blot, the samples can be quantitated using the available software. Excellent quantitation can be obtained with images captured on the Bio-Rad Fluor-S-Max or equivalent instrument using Bio-Rad QuantityOne software. The contour tool is used to select the area for quantitation and the values are background subtracted to give an adjusted volume in counts for each standard and sample. Using above protocol linear standard curves are generated over 1-1.5 orders of magnitude range in target load. It is important to note that immunodetections usually show a strongly sigmoidal signal to load response curve, with a region of trace detection of low loads, a pseudolinear range and a region of saturated response with high loads. For immunoquantitation it is critical that the target proteins in the samples and the standard curve fall within the pseudolinear range. Our total detection range using this protocol spans over 2 orders of magnitude, but the quantifiable range is narrower.
References:
MacKenzie et al (2005). Large reallocations of carbon, nitrogen and photosynthetic reductant among phycobilisomes, photosystems and Rubisco during light acclimation in Synechococcus elongatus are constrained in cells under low environmental inorganic carbon. Arch of Microbiol. 183: 190 - 202.
Bouchard et al. (2006) UVB effects on the photosystem II-D1 protein of phytoplankton and natural phytoplankton communities. Photochem and Photobiol 82: 936-951.
Morash et al. (2007) Macromolecular dynamics of the photosynthetic system over a seasonal developmental progression in Spartina alterniflora. Can J. of Bot. 85: 476-483(8)Application example
.jpg)
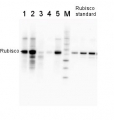
2 µg of total protein from (1-7) Prochlorococcus sp. extracted with Protein Extration Buffer, PEB (AS08 300), (8-11) Rubisco protein standard (AS01 017S) were separated on 4-12% NuPage (Invitrogen) LDS-PAGE and blotted 1h to PVDF. Blots were blocked immediately following transfer for 1h/RT with agitation. Blots were incubated in the primary antibody at a dilution of 1: 20 000 for 1h at room temperature with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, then washed once for 15 min and 3 times for 5 min in TBS-T at room temperature with agitation. Blots were incubated in secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated) diluted to 1:50 000 for 1h/RT with agitation. The blots were washed as above and developed for 5 min with ECL detection reagent according the manufacturers instructions. Images of the blots were obtained using a CCD imager (FluorSMax, Bio-Rad) and Quantity One software (Bio-Rad). There is a low level of a known RbcL cleavage product below the main band in some of the Prochlorococcus samples.
Recommended secondary antibodies: goat anti-rabbit HRP conjugated, goat anti-rabbit ALP conjugated
Recommended chemiluminescent detection reagent: AgriseraECLBrightMethod for embedding in LR WHITE for immunolabelling using anti-RbcL antibody
Prins et al. (2008). Cysteine proteinases regulate chloroplast protein content and composition in tobacco leaves:a model for dynamic interactions with ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) vesicular bodies. J.Ex.Bot.59:1935-1950. Method described below is a courtesy of Dr Aranda
Samples must be sectioned at 1 x 1 mm. 1)Fixation: Glutaraldehyde 0.5% + paraformaldehyde 2.5% in sodium phosphate buffer 0.1M pH= 7.2, 1 hour and 30 min. 2) 3 washes in sodium phosphate buffer 0.1M pH= 7.2 (15 min. each one). 3)Dehydratation and embedding:
-35% Ethanol……30 min.
-50% Ethanol……30 min.
-70% Ethanol……30 min.
-96% Ethanol……30 min.
-100% Ethanol……30 min.
-100% Ethanol……30 min.
-100% Ethanol……30 min.-75% Ethanol + 25% LR White…… 1 hour
-50% Ethanol + 50% LR White…… 1 hour
-25% Ethanol + 75% LR White…… 1 hour
-100% LR White…………………... 1 hour
-100% LR White…………………... overnight.4) Polymerization: Oven 12-18 hours at 50ºC. For flat molds in continous flux of N2
5) Block sectioning: Ultrathin sections (60-70nm) using a ultramicrotome. Ultrathin sections should be placed on formvar coated níkel grids.
IMMUNOLABELLING
1) Blocking: Place grids with sections into 30-μL drop of blocking buffer during 30 minutes. (Blocking buffer: phosphate-buffered saline (PBS) + 5% bovine serum albumin (BSA).
2) Incubation: Incubate in drop of primary antibody, anti-RbcL (Rubisco form I and II, Agrisera) diluted 1:250 in blocking buffer. 3 hours at room temperature (could be extend to overninght).
3) Rinse: Pass the grids through a series of drops of PBS + 1%BSA. Two or three changes during 5 minutes each one.
4) Incubation: Incubate grids in 30-μL of secondary antibody gold-labelled (British BioCell International, 10nm), diluted 1:50 in blocking buffer during 1.5 hours at room temperature.
5) Rinse: Pass the grids through a series of drops of PBS + 1%BSA. Two changes during 5 minutes each one.
6) Rinse: Pass the grids through a series of drops of PBS. Two or three changes during 5 minutes each one.
7) Rinse: Pass the grids through a series of drops of water (ultra-pure water must filtered 0.2 μm) . Four to five changes during 5 minutes each one, allowed to dry for later observation.
8) Staining: The grids may be post-stained with uranyl acetate and lead citrate.
Pao et al. (2018). Lamelloplasts and minichloroplasts in Begoniaceae: iridescence and photosynthetic functioning. J Plant Res. 2018 Mar 2. doi: 10.1007/s10265-018-1020-2. (ImmunoGold)
Liang et al. (2014). Cyanophycin mediates the accumulation and storage of fixed carbon in non-heterocystous filamentous cyanobacteria from coniform mats. PLoS One. 2014 Feb 7;9(2):e88142. doi: 10.1371/journal.pone.0088142. eCollection 2014. (ImmunoGold) - Reviews:
-
Orly Levitan | 2016-09-14I have been using this antibody for the past 7 years. It has worked wonderfully for me every time I tried. I have used it on cyanobacteria (including Trichodesmium), Diatoms, and Seagrasses, with and without the RbcL standard. i have several publication in which I used this antibody (and others by Agrisera). Publication for example: Levitan et al., 2010, Regulation of nitrogen metabolism in the marine diazotroph Trichodesmium IMS101 under varying temperatures and atmospheric CO2 concentrations, Environmental microbiology.Levitan et al., 2015, Remodeling of intermediate metabolism in the diatom Phaeodactylum tricornutum under nitrogen stress, PNASJerzy Kruk | 2014-02-10samples used were thylakoids and leaf powder, 12% SDS gels for PAGE, 5 ug protein per lane or 7 ug/chlorophyll per lane (thylakoids), dilution 1:5000, the observed product at ca. 53 kDa, AB works wellRenata Ditt | 2013-11-26Weve been very happy with this antibody. We have been using it for both westerns and ELISAs, with great results. We work with sea water samples containing a mixed population of species of diatoms and other plankton.Katarzyna Hura | 2013-10-30I am satisfied with this antibody from Agrisera. It gives clean results. We have used this antibody for western blot detection (diluttion 1:20 000).Tanja Göbel | 2012-09-25I´m using the antibodies for the immunodetection on western blots with the fluorescent based licor odyssey system (with the IRDye secondary antibodies Donkey anti rabbit IgG 800CW and 680 (Licor)). I´m analysing whole protein or chloroplasts protein extracts optained from Arabidopsis thaliana. Proteins were either precipitated with TCA/Aceton or directly solubilized with NP-40 in a tris-buffer. The only problem with the unprecipitated samples is that one gets a very strong backround in the 680nm channel due to the chlorophyll-fluorescence. The primary antibodies were diluted in TBS with 0,02% Sodiumazid according to the recommanded dilution (for ECL) in the data sheet and the membranes were incubated for 1 h at RT with agitation. The membranes are blocked with BSA in TBS and washed with TBS-T. I´m reusing the diluted antibodies for up to 5 to 10 western blots without any significant loss of signal (diuluted antibodies are stored at 4°C). The signals I´m getting with the antibodies are very clear and hieghly specific, with only low (anti-Lhcb1 and 2) or nearly no (anti-RbcL and RA) unspecific binding. Usually, I´m loading 10 µg protein and seperate them on 10% Mini-gels. So far I had no problems with Agrisera-antibodies.Sarah Croonenborghs | 2009-05-28The antibody worked excellent on a Guzmania hybrid (Bromeliaceae) in both dilutions, 1: 5000 and 1:10 000.Enrique Olmos Aranda. CEBAS-CSIC. Spain | 2009-02-20We have satisfactorily used this antibody with Paspalum and tobacco for immunolabelling.Dilution: 1/250