1
Anti-Cat | Catalase (peroxisomal marker)
AS09 501 | Clonality: Polyclonal | Host: Rabbit | Reactivity: A.thaliana, A.madagascariensis, B. distachyon, B.oleracea, Cirtus sp., Fragaria x ananassa, H.vulgare, L. sativus, L. albus, M. perniciosa, N.bentamina, N.tabacum, O.sativa, P. kurroa, P.sativum, P. zeylandica, P.tomentosa, R.sativus, S. italica, S.lycopersicum, S.oleracea, T.aestivum, Z.mays, V. vinifera
Benefits of using this antibody
- Product Info
-
Immunogen: KLH-conjugated peptide chosen from know plant catalase sequences including Arabidopsis thaliana isoforms: catalase-1 (Q96528, At1g20630), catalase-2 (P25819, At4g35090), catalase-3 (Q42547, At1g20620);
Host: Rabbit Clonality: Polyclonal Purity: Serum Format: Lyophilized Quantity: 50 µl Reconstitution: For reconstitution add 50 µl of sterile water Storage: Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please remember to spin the tubes briefly prior to opening them to avoid any losses that might occur from material adhering to the cap or sides of the tube. Tested applications: Immunogold (IG), Immunolocalization (IL), Immunoprecipitation (IP), Western blot (WB) Recommended dilution: 1: 500 (IG), 1: 25 - 1: 500 (IL), 2 µg (IP), 1 : 1000 (WB) Expected | apparent MW: 57 | 55 kDa
- Reactivity
-
Confirmed reactivity: Arabidopsis thaliana, Aponogeton madagascariensis, Brachypodium distachyon, Brassica napus, Brassica oleracea, Fragaria x ananassa, Fragaria x ananassa, Hordeum vulgare, Lathyrus sativus, Lupinus albus, Lupinus luteus, Moniliophthora perniciosa, Musa acuminate, Musa paradisiaca L., Nicotiana bentamina, Nicotiana tabacum, Oryza sativa, Paulownia tomentosa, Picrorhiza kurroa, Pisum sativum, Plumbago zeylanica, Setaria italica L. P. Beauv, Raphanus sativus, Solanum lycopersicum, Spinacia oleracea, Triticum aestivum, Zea mays, Vitis vinifera Predicted reactivity: Avicennia marina, Betula pendula, Brassica campestris, Brassica napus, Brassica rapa subsp. pekinensis, Citrus sp., Citrus clementina, Citrus maxima, Camellia sinensis, Cucumis sativus, Cucurbita maxima, Cucurbita moschata, Elaeis guineensis var. tenera, Eucalyptus grandis, Glycine max, Gossypium mexicanum, Helianthus annus, Hibiscus cannabinus, Litchi chinensis, Lupinus albus, Malus domestica, Manihot esculenta, Marchantia polymorpha, Morus notabilis, Nicotiana tabacum, Paenibacillus sp., Pinus pinea, Populus albaxtremula, Populus jackii, Prunus persica, Raphanus sativus, Saccharum officinarum, Sesamum indicum, Solanum tuberosum, Ulva prolifera, Vicia faba
Species of your interest not listed? Contact usNot reactive in: Chlamydomonas reinhardtii - Application Examples
-
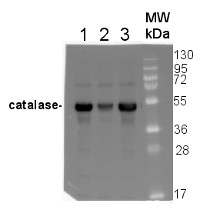
10 µg of total protein from Arabidopsis thaliana Col0 (1), Cat2-(Col0) (2), Ler0 (3), Cat2-(Ler0) (4), Zea mays (5), Oryza sativa (6), Brassica oleracea (7), Nicotiana bentamina (8) were extracted with 60mM Tris pH 6.9, 10mM DTT, 20% glycerol, 1mm PMSF were separated on 12.5% SDS-PAGE and blotted 1h to PVDF. Blot was blocked with 3% skim milk in PBS+0.05% Tween20 for 1h at room temperature (RT) with agitation. Blot was incubated in the primary antibody at a dilution of 1: 1 000 for 1h at RT with agitation in the same buffer. The antibody solution was decanted and the blot was rinsed briefly three times, then washed 3 times for 5 min in PBS-T at RT with agitation. Blot was incubated in secondary antibody (anti-rabbit IgG horse radish peroxidase conjugated, Agrisera, AS09 602) diluted to 1:50 000 in 3% skim milk in PBS+0.05% Tween20 for 1h at RT with agitation. The blot was washed as above and developed for 1 min with Western Lightning Plus-ECL ( PerkinElmer )according to the manufacturers instructions. Exposure time was 5min. in ChemiDoc XRS+ (Biorad ).
Courtesy of Brigitte van de Cotte, Gent University, Belgium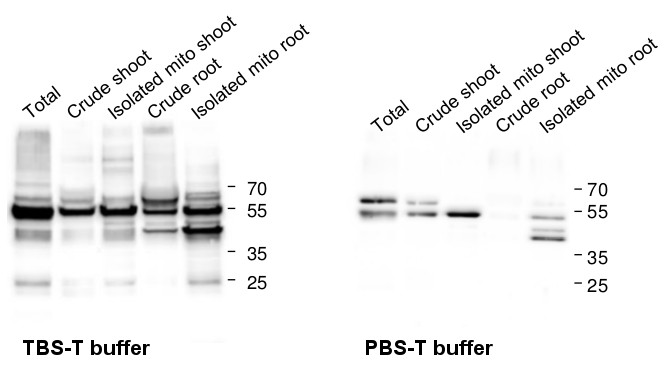
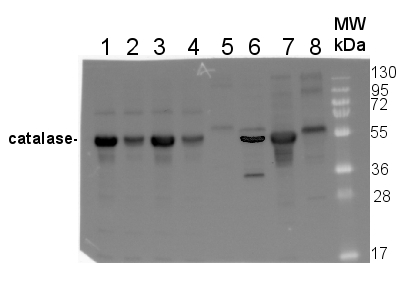
Blots were performed from 10 µg of protein from total extracts, crude extracts as well as from isolated tagged-mitochondria (leaves or roots). Arabidopsis thaliana protein extracts were prepared using a protein extraction buffer (100 mM Tris-HCl pH 7.5, 50 mM EDTA, 250 mM NaCl, 0.05% SDS). Samples were denatured with Laemmli buffer (Bio-Rad) supplemented with 10% β-mercaptoethanol at 95°C for 10 min before separating the protein mixtures on reducing 12% polyacrylamide gel. Protein extracts were blotted 1h onto a 0.45 µm nitrocellulose membrane using wet transfer. Blots were blocked with 5% milk for 1h/RT with agitation. Blots were incubated with the primary antibodies anti-catalase (Agrisera, AS09 501) at a dilution of 1: 1 000 for ON/4°C with agitation in TBS-T or PBS-T + 2% milk, respectively. Blots were washed 3 times for 5 min in TBS-T or PBS-T at RT with agitation. Blot was incubated for 1h/RT with agitation in Agrisera matching secondary antibodies (anti-rabbit IgG horse radish peroxidase conjugated, AS09 602) diluted to 1:10 000 in TBS-T or PBS-T + 2% milk. Blots were washed 3 times for 5 min in TBS-T or PBS-T following by 3 additional washing steps for 5 min in TBS or PBS. Visualization was carried out using the chemiluminescence kit Agrisera ECLBright; AS16 ECL-N-100 and signals were detected using Azure c600 Western Blot Imaging system (Azure biosystems). Exposure time was 2-5 min.
Courtesy of Dr Jonathan Przybyla-Toscano, Umeå Plant Science Centre, SwedenApplication examples: 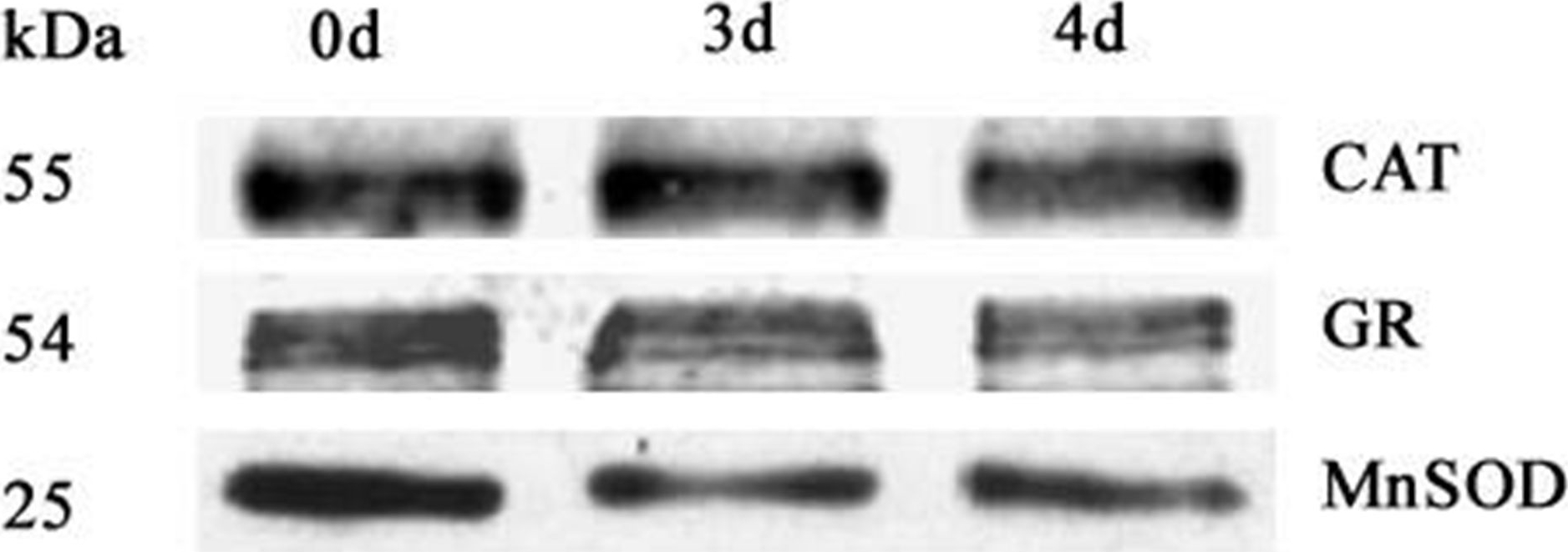
Reactant: Oryza sativa (Asian rice)
Application: Western Blotting
Pudmed ID: 27124767
Journal: PLoS One
Figure Number: 7A
Published Date: 2016-04-29
First Author: Yin, G., Whelan, J., et al.
Impact Factor: 2.942
Open PublicationWestern blot analysis of antioxidant enzymes in purified mitochondria from 0 d, 3 d, and 4 d aged seeds.Total 10 ?g protein was separated by SDS gel electrophoresis and blotted to supported polyvinylidene difluoride, then probed with antibodies against catalase (CAT), glutathione reductase (GR) and manganese superoxide dismutase (MnSOD).
Reactant: Plant
Application: Western Blotting
Pudmed ID: 28389868
Journal: Planta
Figure Number: 6A
Published Date: 2017-07-01
First Author: Dauphinee, A. N., Fletcher, J. I., et al.
Impact Factor: 3.95
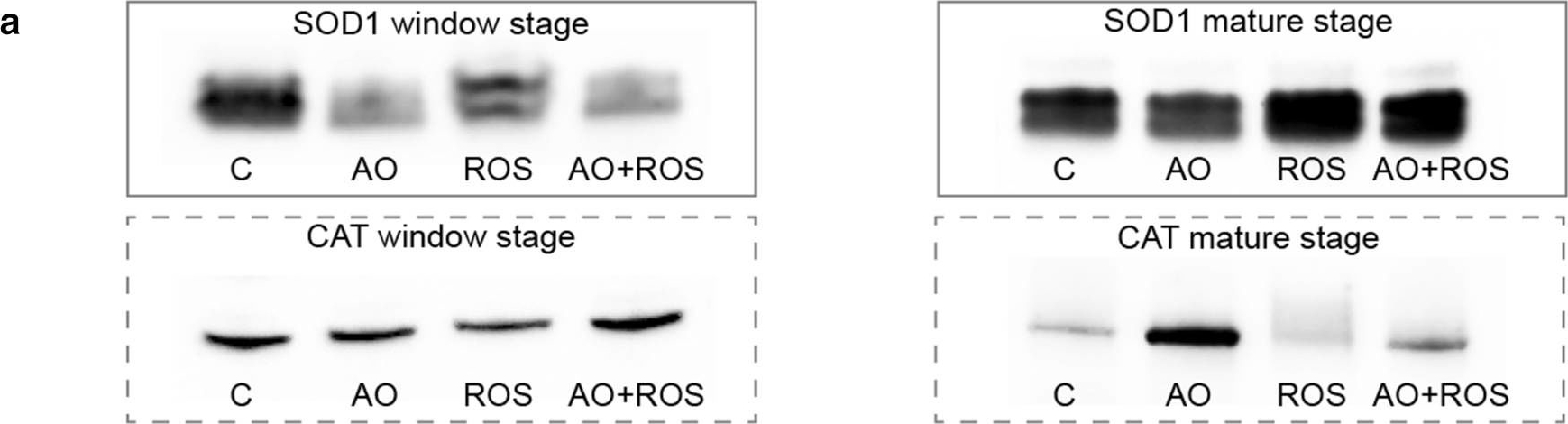
Open PublicationDetection of superoxide dismutase 1 (SOD1) and catalase (CAT) using western blotting for the control (C), antioxidant (AO; 400 µg/ml ascorbic acid and 200 µg/ml cysteine), 1 mM H2O2 reactive oxygen species (ROS), and AO + ROS treatment groups. Immunoprobing for SOD1 and CAT (a). SOD1 mean protein band intensities for window stage (b) and mature leaves (c). CAT mean protein band intensities for window stage (d) and mature leaves (e). One-way ANOVA, Dunnett’s multiple comparison test, **P < 0.01; *P < 0.05 ns, non-significant; n ? 4. Error bars represent standard error
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 28391398
Journal: Plant Mol Biol
Figure Number: 3C
Published Date: 2017-05-01
First Author: Bi, C., Ma, Y., et al.
Impact Factor: 3.808
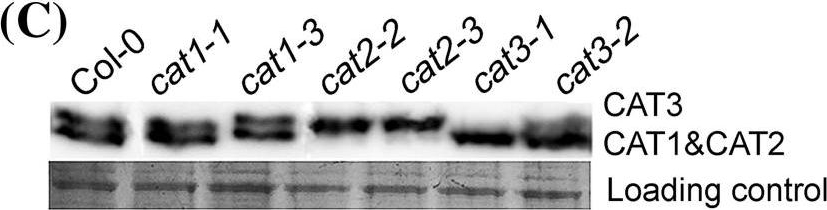
Open PublicationMolecular and biochemical characterizations of the catalase mutants. a Structures of the CAT1, CAT2 and CAT3 genes are shown with the T-DNA insertion sites in cat1-1 (SAIL_525_C10), cat1-3 (SALK_208924), cat2-2 (SALK_057998), cat2-3 (SALK_144919), cat3-1 (SALK_092911) and cat3-2 (SALK_088601). Exons are indicated by black blocks, introns by solid lines and untranslated regions by grey broken lines. The black arrowhead indicates the orientation of the T-DNA insertion. b Quantitative RT-PCR analysis of CAT gene expressions in wild-type and catalase mutants is shown. Each value for real-time PCR is the mean?±?SE of three independent experiments. c Protein gel blot analysis was performed using anti-catalase serum and total proteins were extracted from seeds grown under 16-h light/8-h dark conditions for 24 h after stratification. Protein extracts were stained with Ponceau S as a loading control. The protein gel blot assay was repeated independently three times. d The effect of 3-AT on seed germination in catalase mutants. Germination rates were recorded for the wild-type, CAT homozygous mutants grown under light conditions (16 h light/8 h dark) for 60 h after stratification on MS medium supplemented with 0, 5mM 3-AT. Each value is the mean?±?SE of three independent biological experiments, and different letters indicate significant differences at P?
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 28391398
Journal: Plant Mol Biol
Figure Number: 4D
Published Date: 2017-05-01
First Author: Bi, C., Ma, Y., et al.
Impact Factor: 3.808
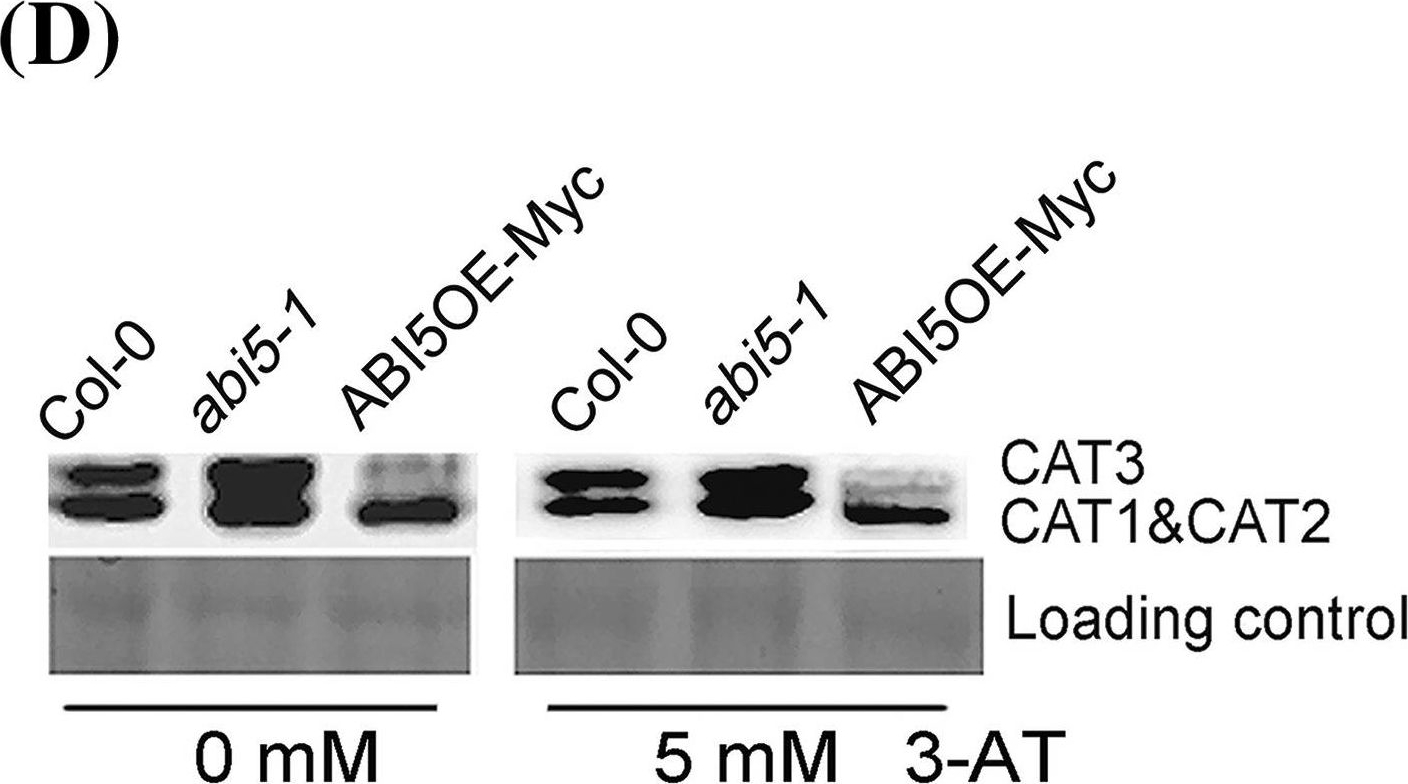
Open PublicationExpression analysis of CAT members in the seeds of Col-0, ABI5OE-Myc and abi5-1. The seeds of Col-0, ABI5OE-Myc and abi5-1 were grown under 16-h light/8-h dark conditions for 24 h after stratification on MS medium supplemented with 0 or 5mM 3-AT. a–c are quantitative RT-PCR analysis of CAT1 (A), CAT2 (B) and CAT3 (C), respectively. Each value is the mean?±?SE of three independent experiments. d Protein gel blot analysis of the expression of CAT in the seeds of Col-0, ABI5OE-Myc and abi5-1. Protein extracts were stained with the Ponceau S as a loading control. The assay was independently repeated three times
Reactant: Moniliophthora perniciosa (Witches' broom disease)
Application: Western Blotting
Pudmed ID: 28818052
Journal: BMC Microbiol
Figure Number: 4A
Published Date: 2017-08-17
First Author: Mares, J. H., Gramacho, K. P., et al.
Impact Factor: 3.437
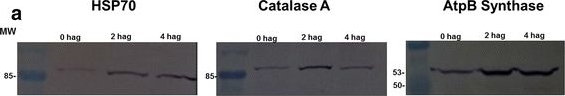
Open PublicationAccumulation of BiP, Catalase, and ATP synthase (beta chain) by western blot and analysis of spots. a Image of a nitrocellulose membrane hybridized with Anti-BiP, Anti-Catalase, and Anti-ATP synthase (beta chain) of Arabidopsis (Normalization, see Additional file 6: Figure S4). b Relative accumulation determined from images of the nitrocellulose membrane, using the Gel Quant. Net 1.8.0 software. c Relative quantification of each spot corresponding to proteins analyzed by western blot. The average values of the each spot’s percentage volume in each gel replicate was used for graph construction

Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 30609769
Journal: Int J Mol Sci
Figure Number: 3B
Published Date: 2019-01-02
First Author: Szyma?ska, K. P., Polkowska-Kowalczyk, L., et al.
Impact Factor: 5.542
Open PublicationSnRK2.4 and SnRK2.10 modulate catalase (CAT) on multiple levels during response to salt stress. Wild type and snrk2 mutants’ seedlings were subjected to treatment with 150 mM NaCl for times indicated. CAT1 expression was determined by RT-qPCR (A), total catalase protein was determined by immunoblot analysis (B), and total catalase activity assay was performed (C); error bars represent SD; stars represent statistically significant differences in comparison with the wt plants (Student t-test) where * p < 0.05; ** p < 0.001; *** p < 0.0001. After exposure, membranes were stripped and reused for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) detection as a loading control. At least two independent biological replicates of the experiment were performed, each with four samples per data point. Results of one representative experiment are shown.
Reactant: Arabidopsis thaliana (Thale cress)
Application: Western Blotting
Pudmed ID: 33445496
Journal: Plants (Basel)
Figure Number: 4C,D
Published Date: 2021-01-12
First Author: Dmitrovi?, S., Dragi?evi?, M. B., et al.
Impact Factor: None
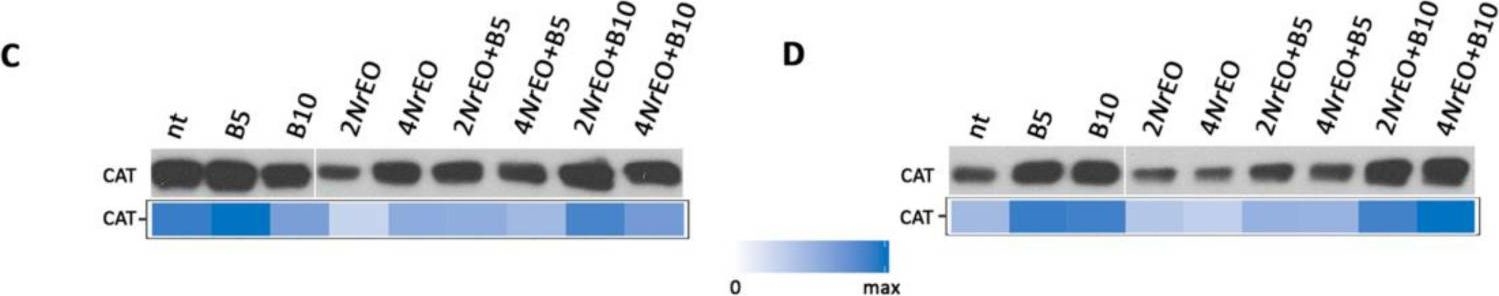
Open PublicationCatalases (CAT) activity in Arabidopsis grown in vitro, measured 10 days after treatment with BASTA applied at two concentrations B5 (5 mg L?1) and B10 (10 mg L?1), N. rtanjensis essential oil applied at two concentrations 2% (2NrEO) and 4% (4NrEO), and of their combinations. (A,B) CAT activity stain after native-PAGE separation (5 µg per lane of total protein), in shoots and roots, respectively. The detected activities were measured densitometrically and presented as relative CAT activity (% of control) in comparison to non-treated plants (nt). (C,D) CAT immunoblot (20 µg of soluble proteins per line) in shoots and roots, respectively. Heat-maps show relative abundances of CAT proteins. Maximal values on the color scales represent maximal values recorded for each immunoblot.
- Additional Information
-
Additional information: This antibody is recognizing all three isoforms of Arabidopsis thaliana catalase. Catalase-2 is a main isoform expressed in leaf tissue and localized to peroxisomes.
This antibody contains 0.1 % ProClin.Additional information (application): To obtain reactivity with Solanum lycopersicum urea gel needs to be apply. Please, contact us for more details.
To decrease background signal this antibody needs to be incubated in PBS-T NOT TBS-T. For reference, check image in application example below. - Background
-
Background: Catalase is an enzyme found in most living organisms which is catalazying decomposition of hydrogen peroxide to water and oxygen. In plant cells catalase is found in peroxisomes. This enzyme is involved in photorespiration and symbiotic nitrogen fixation.
- Product Citations
-
Selected references: Piechowiak et al. (2025), Ozone Treatment Enhances Antioxidant Status and Energy Metabolism in Radish (Raphanus sativus) Sprouts. Springer Nature, Food Bioprocess Technology.
Krupinska et al. (2025). Iron allocation to chloroplast proteins depends on the DNA-binding protein WHIRLY1. Planta. 2025 Jun 17;262(2):32. doi: 10.1007/s00425-025-04736-8.
Boussardon et al. (2025). The atypical proteome of mitochondria from mature pollen grains. Curr Biol . 2025 Jan 21:S0960-9822(24)01705-6. doi: 10.1016/j.cub.2024.12.037. Niemiro et al. (2024). LSU family members and NBR1 are novel factors that contribute to homeostasis of catalases and peroxisomes in Arabidopsis thaliana. Sci Rep. 2024 Oct 25;14(1):25412. doi: 10.1038/s41598-024-76862-4.
Hu et al. (2023).Catalase associated with antagonistic changes of abscisic acid and gibberellin response, biosynthesis and catabolism is involved in eugenol-inhibited seed. Plant Cell Rep. 2023 Dec 23;43(1):10.doi: 10.1007/s00299-023-03096-5.
Melicher et al. (2023). Methyl viologen-induced changes in the Arabidopsis proteome implicate PATELLIN 4 in oxidative stress responses.J Exp Bot. 2024 Jan 1;75(1):405-421.doi: 10.1093/jxb/erad363.
Boussardon C, Carrie C, Keech O. Comparing plastid proteomes points towards a higher plastidial redox turnover in vascular tissues than in mesophyll cells. J Exp Bot. 2023 Apr 7:erad133. doi: 10.1093/jxb/erad133.
Cembrowska-Lech, Rybak (2023) Nanopriming of Barley Seeds-A Shotgun Approach to Improve Germination under Salt Stress Conditions by Regulating of Reactive Oxygen Species. Plants (Basel). 2023;12(2):405. Published 2023 Jan 15. doi:10.3390/plants12020405
Fortunato et al. (2022) GUN1 involvement in the redox changes occurring during biogenic retrograde signaling. Plant Science. Volume 320, July 2022, 111265.
Vinegra de la Torre et al. (2022). FLOWERING REPRESSOR AAA+ ATPase 1 is a novel regulator of perennial flowering in Arabis alpina. New Phytol. 2022 Oct;236(2):729-744. doi: 10.1111/nph.18374. Epub 2022 Aug 8. PMID: 35832005.(Immunogold)
Tokarz et al. (2021). Stem Photosynthesis-A Key Element of Grass Pea (Lathyrus sativus L.) Acclimatisation to Salinity. Int J Mol Sci. 2021 Jan 12;22(2):685. doi: 10.3390/ijms22020685. PMID: 33445673; PMCID: PMC7828162.
Li et al. (2021) Isolation and comparative proteomic analysis of mitochondria from the pulp of ripening citrus fruit. Hortic Res. 2021 Feb 1;8(1):31. doi: 10.1038/s41438-021-00470-w. PMID: 33518707; PMCID: PMC7848011.
Wilmowicz et al (2021) EPIP-Evoked Modifications of Redox, Lipid, and Pectin Homeostasis in the Abscission Zone of Lupine Flowers. International Journal of Molecular Sciences. 2021; 22(6):3001. https://doi.org/10.3390/ijms22063001
Bapatla et al. (2021). Modulation of Photorespiratory Enzymes by Oxidative and Photo-Oxidative Stress Induced by Menadione in Leaves of Pea (Pisum sativum). Plants 10, no. 5: 987. https://doi.org/10.3390/plants10050987
Adamiec et al. (2021). Fatty acid composition and cpDNA content in Arabidopsis thaliana mutants deprived of EGY1 protease. PHOTOSYNTHETICA 59 (4): 633-639, 2021, DOI 10.32615/ps.2021.053
Santos et al.(2020). Diversity of banana diploid hybrids: An assessment based on a hydroponic system. Plant Breeding Volume 139, Issue 6. doi: 10.1111/pbr.12869
Tokarz et al. (2020). Can Ceylon Leadwort ( Plumbago zeylanica L.) Acclimate to Lead Toxicity?-Studies of Photosynthetic Apparatus Efficiency. Int J Mol Sci. 2020 Mar 9;21(5):1866.doi: 10.3390/ijms21051866.
Boussardon et al. (2020). Tissue-specific Isolation of Arabidopsis/plant Mitochondria - IMTACT (Isolation of Mitochondria Tagged in Specific Cell Types). Plant J. 2020 Feb 14. doi: 10.1111/tpj.14723. Online ahead of print.
Rodriguez et al. (2020). Autophagy mediates temporary reprogramming and dedifferentiation in plant somatic cells. bioRxiv doi.org/10.1101/747410
Calero-Munoz et al. (2019). Cadmium induces reactive oxygen species-dependent pexophagy in Arabidopsis leaves. Plant Cell Environ. 2019 Sep;42(9):2696-2714. doi: 10.1111/pce.13597.
Mares et al. (2020). Hydrosoluble phylloplane components of Theobroma cacao modulate the metabolism of Moniliophthora perniciosa spores during germination.Fungal Biol. 2020 Jan;124(1):73-81. doi: 10.1016/j.funbio.2019.11.008.
Calero-Munoz et al. (2019). Cadmium induces ROS-dependent pexophagy in Arabidopsis leaves. Plant Cell Environ. 2019 May 31. doi: 10.1111/pce.13597.
Szymanska et al. (2019). SNF1-Related Protein Kinases SnRK2.4 and SnRK2.10 Modulate ROS Homeostasis in Plant Response to Salt Stress. Int J Mol Sci. 2019 Jan 2;20(1). pii: E143. doi: 10.3390/ijms20010143.
Bastow et al. (2018). Vacuolar Iron Stores Gated by NRAMP3 and NRAMP4 Are the Primary Source of Iron in Germinating Seeds. Plant Physiol. 2018 Jul;177(3):1267-1276. doi: 10.1104/pp.18.00478.
Pan et al. (2018). Comparative proteomic investigation of drought responses in foxtail millet. BMC Plant Biol. 2018 Nov 29;18(1):315. doi: 10.1186/s12870-018-1533-9.
Su et al. (2018). The Arabidopsis catalase triple mutant reveals important roles of catalases and peroxisome derived signaling in plant development. J Integr Plant Biol. 2018 Mar 25. doi: 10.1111/jipb.12649.
Kang et al. (2018). Autophagy-related (ATG) 11, ATG9 and the phosphatidylinositol 3-kinase control ATG2-mediated formation of autophagosomes in Arabidopsis. Plant Cell Rep. 2018 Jan 19. doi: 10.1007/s00299-018-2258-9.
Mares et al. (2017). Proteomic analysis during of spore germination of Moniliophthora perniciosa, the causal agent of witches' broom disease in cacao. BMC Microbiol. 2017 Aug 17;17(1):176. doi: 10.1186/s12866-017-1085-4.
Sultan et al. (2017). The Reverse Transcriptase/RNA Maturase Protein MatR Is Required for the Splicing of Various Group II Introns in Brassicaceae Mitochondria. Plant Cell. 2016 Nov;28(11):2805-2829.
Zhang et al. (2017). Global analysis of protein lysine succinylation profiles in common wheat. BMC Genomics. 2017 Apr 20;18(1):309. doi: 10.1186/s12864-017-3698-2. (Triticum aestivum L., immunoprecipitation)
Kneeshaw et al. (2017). Nucleoredoxin guards against oxidative stress by protecting antioxidant enzymes. Proc Natl Acad Sci U S A. 2017 Jul 19. pii: 201703344. doi: 10.1073/pnas.1703344114.
Dauphinee et al. (2017). Remodelling of lace plant leaves: antioxidants and ROS are key regulators of programmed cell death. Planta. 2017 Apr 7. doi: 10.1007/s00425-017-2683-y. (Aponogeton madagascariensis)
Yin et al. (2016). Comprehensive Mitochondrial Metabolic Shift during the Critical Node of Seed Ageing in Rice. PLoS One. 2016 Apr 28;11(4):e0148013. doi: 10.1371/journal.pone.0148013. eCollection 2016.
Lee et al. (2016). Superoxide serves as a putative signal molecule for plant cell division: overexpression of CaRLK1 promotes the plant cell cycle via accumulation of O2 − and decrease in H2O2. Physiol Plantarum. doi:10.1111/ppl.12487
Piechowiak et al (2023) Effect of postharvest nicotinamide treatment on NAD+ metabolism and redox status in strawberry fruit during storagev - Protocols
-
Agrisera Western Blot protocol and video tutorials
Protocols to work with plant and algal protein extracts
Oxygenic photosynthesis poster by prof. Govindjee and Dr. Shevela
Z-scheme of photosynthetic electron transport by prof. Govindjee and Dr. Björn and Dr. Shevela - Reviews:
-
Xiaohong | 2024-09-05This antibody worked quite well with clean background for samples obtained from Arabidopsis seedlings.?ukasz W. | 2020-11-27We have used this Ab in our lab for several years. We work on several lupinus species like L. luteus, L. albus. We tested this Ab also on Brassica napus. It works well, the reactivity is good, the bands are well visible, and backgroud is very week. It is worth to recommend.catalase | 2020-09-17Very good antibody. Using Arabidopsis seedlings for protein extraction. The bands are clearly at 55kDa without any strong noisy bands.
Highly recommended.Arek | 2018-10-22This product was proved to work perfectly with Lolium/Festuca species. Thus, it seems that it has a wide range of species compatibility.Adrian D. | 2016-07-27The antibody worked very well for protein extractions (1-20µg protein loaded) from Aponogeton madagascariensis leaves.