| Calculated and apparent molecular weight can differ by 10-20 kDa, therefore careful analysis of obtained band pattern, combined with reliable molecular weight markers is helpful to secure correct interpretation of obtained results. There are several reasons which may play a role. To name a few:
To check if a target protein is subjected to PTMs following approaches can be applied:
| 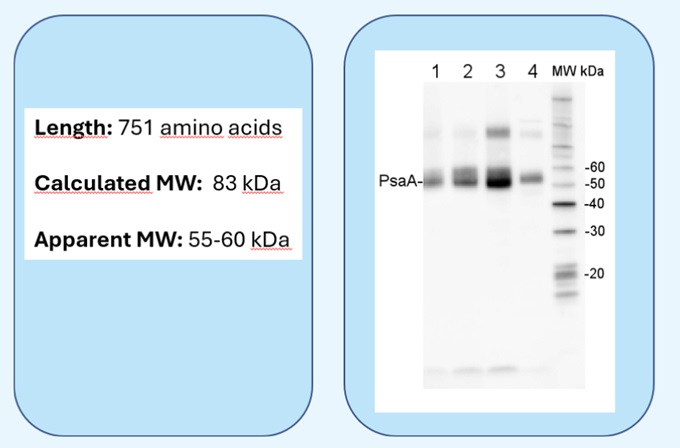 Example: detection of a photosynthetic protein, PsaA, using Agrisera antibody AS06 172. |
Latest
Calclulated and aparent molecular weight of detected protein is different, why?2025-10-03 Is an antibody going to work in a technique I am planning to use it in?
2025-09-30 Antibody reactivity to recombinant protein,does not validate antibody specificity in endogenous sample
2025-08-20 Can aggregated antibody be still used?
2025-08-15 Blot Once, Probe Twice - when such approach can be beneficial?
2025-07-31 Why should glycerol not be added, to stabilize antibodies in serum?
2025-06-27 Can methanol in transfer buffer be replaced by a less toxic alternative?
2025-05-12 Will the precipitation of SDS in the stripping buffer affect antibody stripping efficiency?
2025-04-29 How to check if protein transfer from a gel to a membrane was efficient
2025-04-15 How to detect membrane proteins on Western blot?
2025-03-27
Archive
- October - 2025
- September - 2025
- August - 2025
- July - 2025
- June - 2025
- May - 2025
- April - 2025
- March - 2025
- February - 2025
- January - 2025
- December - 2024
- November - 2024
- September - 2024
- July - 2024
- June - 2024
- May - 2024
- March - 2024
- February - 2024
- December - 2023
- November - 2023
- September - 2023
- July - 2023
- May - 2023
- March - 2023
- January - 2023
- December - 2022
- November - 2022
- October - 2022
- September - 2022
- August - 2022
- June - 2022
- May - 2022
- March - 2022
- February - 2022
- January - 2022
- November - 2021
- October - 2021
- August - 2021
- June - 2021
- May - 2021
- April - 2021
- March - 2021
- February - 2021
- January - 2021
- December - 2020
- November - 2020
- October - 2020
- September - 2020
- August - 2020
- July - 2020
- June - 2020
- May - 2020
- April - 2020
- March - 2020
- January - 2020
- November - 2019
- October - 2019
- March - 2019
- April - 2017
- February - 2017
- May - 2016
- February - 2014
- September - 2013
- December - 2010
